
DESIGN OF A STANDOFF OBJECT-ORIENTED MARKUP
LANGUAGE (SOOML) FOR ANNOTATING
BIOMEDICAL LITERATURE
Jing Ding and Daniel Berleant
Department of Electrical & Computer Engineering, Iowa State University, Ames, IA, USA
Keywords: text mining, markup language, bioinformatics
Abstract: With the rapid growth of electronically available scientific literature, text mining is attracting increasing
attention. While numerous algorithms, tools, and systems have been developed for extracting information
from text, little effort has been focused on how to mark up the information. We present the design of a
standoff, object-oriented markup language (called SOOML), which is simple, expressive, flexible, and ex-
tensible, satisfying the demanding needs of biomedical text mining.
1 INTRODUCTION
With the rapid growth of electronically available
biomedical literature, information extraction from
unrestricted text is attracting increasing attention.
While numerous algorithms and tools have been
developed for extracting the information, little effort
has been focused on markup methodology, i.e., how
to annotate and manage the extracted information.
Among the most intuitive solutions are inline XML
1
markup, as in the GENIA corpus (Kim et al., 2003)
or the Medstract Gold Standards Corpora
2
. Although
straightforward, inline markup is not sufficient for
dealing with complex annotation structures, nor does
it cope well with other issues such as copyright and
annotation reuse, etc. We propose a standoff object-
oriented markup language (SOOML) and present its
design. The design is based on software engineering
principles and intended to meet the challenging
needs of text mining annotations.
In section 2, we analyze the requirements of text
mining annotations and review existing markup
methodologies. Section 3 describes the concepts and
mechanisms in the design of SOOML. A brief dis-
cussion of how the design meets the requirements is
presented in section 4.
1
http://www.w3.org/XML
2
http://medstract.org/gold-standards.html
2 MOTIVATION OF THE WORK
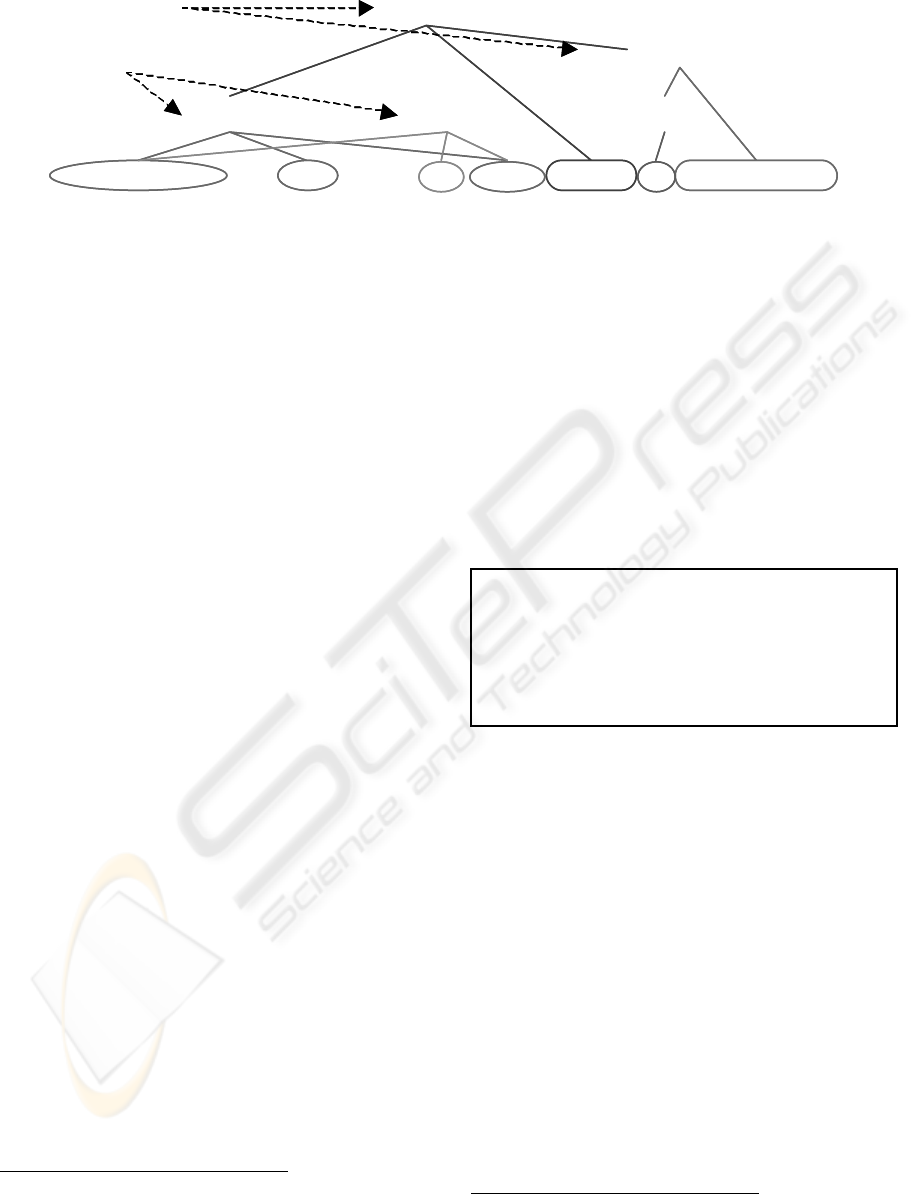
Annotation structures in bioinformatics text mining
can be complex, as illustrated in Fig. 1. In the figure,
we wish to annotate three entities, protein kinase C
alpha isoform, protein kinase C beta isoform and Akt.
The surface strings of the first two entities not only
contain gaps, but also overlap with each other. There
are also two events in the sentence fragment. The
events and the entities have hierarchical relation-
ships. For instance,
entity[3] plays a role in event[a];
and
event[a] itself plays a role in event[b]. This com-
plexity poses demanding requirements for a markup
language.
2.1 Annotation requirements
Expressiveness: Complex annotation structures
(gaps, overlaps, and hierarchies, etc.) can be ex-
pressed easily and intuitively.
Resolution: In many text-mining applications, it
is important to show where the extracted information
is in the original text. The required resolution varies.
Reusability: The language must be able to sup-
port annotation reuse–i.e., annotating annotated text.
Independence of availability: The availability
of annotations should not be restricted by the copy-
rights on the original documents.
382
Ding J. and Berleant D. (2005).
DESIGN OF A STANDOFF OBJECT-ORIENTED MARKUP LANGUAGE (SOOML) FOR ANNOTATING BIOMEDICAL LITERATURE.
In Proceedings of the Seventh International Conference on Enterprise Information Systems, pages 382-385
DOI: 10.5220/0002509603820385
Copyright
c
SciTePress
Flexibility: The language should adapt well to
various text formats and annotation needs.
Efficiency: It should be time and space efficient
in storing, retrieving and processing annotations.
Extensibility: The language should be easily ex-
tensible to incorporate biomedical ontologies (e.g.,
UMLS
3
, and Gene Ontology
4
).
2.2 Existing works
There are some initiatives to develop “standard”
annotated corpora in the bioinformatics field, nota-
bly the GENIA corpus and the Medstract Gold Stan-
dards Corpora. Both annotate MEDLINE
5
abstracts
using in-line XML tags. In-line markup has some
limitations:
• Gapped, overlapped or hierarchical structures
cannot be easily expressed.
• Availability of the annotations may be restricted
by the copyright of the original document.
• In-line markup “contaminates” the original docu-
ments. The “contamination” complicates pipelined
processing, because later stages cannot see di-
rectly the original text.
A solution to these limitations is standoff markup
(annotations stored separately from the original
documents). With the standoff approach, the ques-
tion arises as to how to map an annotation back to its
original text. The annotation graph and W3C’s
XPointer framework address the issue.
The annotation graph (AG) is an abstraction of
existing standoff annotation formats used by linguis-
tic databases for textual and audio signals (Bird and
Liberman, 1999). The signals are viewed as one-
dimensional streams. An AG is a directed graph
with nodes representing time points or character
positions in a stream and with edges representing the
text between two positions or audio content between
two time points. Annotations are labeled on the
edges. Conceptually, AG is equivalent to inline
XML, with nodes corresponding to start and end
3
http://www.nlm.nih.gov/research/umls/
4
http://www.geneontology.org/
5
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi
tags, and edges the text enclosed in tag pairs. Hence,
AG has the same limited expressive power as inline
XML.
The XPointer framework
6
is a W3C standard for
identifying text fragments in XML files. Two
mechanisms (XPath
7
and string matching) are used
in XPointer for different resolutions. XPath is for
identifying text nodes (text fragments, such as titles
or abstracts in MEDLINE files). The string-
matching mechanism is for locating exact words
within a node. Together, they provide the resolution
required by SOOML. However, XPointer has draw-
backs. First, XPointer expressions can be long and
complex. For example, the XPointer expression for
entity[1] in Fig. 1 would be something like this:
More than 200 characters are needed to describe the
original string of 29 characters. Another drawback is
that ambiguity may arise if the matching string oc-
curs multiple times in a text node. To avoid the am-
biguity, the expression has to be longer and more
complex. This complexity and inefficiency make
XPointer not an ideal match to the requirements.
3 DESIGN
Since none of the existing systems mentioned above
fully meets the requirements, we therefore designed
a standoff object-oriented markup language
(SOOML), which defines one concept (monad) and
four mechanisms (object orientation, standoff
markup, annotation mapping, and annotation inclu-
sion). Please keep in mind that SOOML is not an-
other new member of the ever-growing xxML fam-
6
http://www.w3.org/TR/xptr-framework/
7
http://www.w3.org/TR/xpath
Figure 1: Sample annotations in biomedical domain.
… protein kinase C (PKC) alpha, but not beta, isoform inhibited Akt phosphorylation…
entity[1] entity[2]
event[a]
event[b]
entity[3]
Gap/Overlap
Hierarchy
… protein kinase C (PKC) alpha, but not beta, isoform inhibited Akt phosphorylation…
entity[1] entity[2]
event[a]
event[b]
entity[3]
Gap/Overlap
Hierarchy
xpointer(
string-range(//MedlineCitation[PMID=1234]//AbstractText,
‘protein kinase C’) |
string-range(//MedlineCitation[PMID=1234]//AbstractText,
‘alpha’) |
string-range(//MedlineCitation[PMID=1234]//AbstractText,
‘isoform’))
DESIGN OF A STANDOFF OBJECT-ORIENTED MARKUP LANGUAGE (SOOML) FOR ANNOTATING
BIOMEDICAL LITERATURE
383

ily, such as GPML (the schema used in the GENIA
corpus) or SBML (Systems Biology Markup Lan-
guage) (Hucka et al., 2003). A typical xxML de-
fines a set of XML tags (a small ontology) for a spe-
cific purpose, usually focusing on WHAT is marked
up. SOOML, on the other hand, is more interested
in WHERE is the markup. Another important dif-
ference is the relationship with XML. While those
xxMLs are all extensions to XML, SOOML has no
direct connection with XML. It is a set of concepts
and mechanisms, which can be implemented in
XML syntax, C/Java-style syntax, or any other cus-
tom-made syntax. For simplicity, we will use Java-
style syntax to show examples in the following sec-
tions.
3.1 The monad concept
The monad concept as used here is a variant of the
concept with the same name in the text database
field (Doedens, 1994). A stream of text is broken
down into atomic units (tokens) at specified delimit-
ing characters (e.g., the space character). Integer
indexes are assigned to each token in the order of
text-flow. A monad is a token plus its assigned in-
dex. Delimiters may be counted as monads or dis-
carded. If no delimiter is specified, each character is
a monad. Fig. 2 shows a list of monads tokenized at
the space character with the delimiters discarded.
The context of a monad is the underlying text
stream (the text context) along with the delimiting
character set and whether delimiters are included or
discarded (the delimiting context). Thus, given a
context, an integer is unambiguously mapped to a
text token, and vice versa.
3.2 Mechanisms
Object orientation. All annotations are instances of
various classes. All classes are directly or indirectly
derived from the base class
annotation (Inheritance).
An
annotation object may contain other objects as
components (composition) and is responsible for
storing the extracted information (data abstraction
and encapsulation).
Standoff. A collection of various objects is
stored in an annotation file separate from the origi-
nal document. The objects are identified by their
class types and unique IDs.
Annotation mapping. Annotation mapping is
achieved at three levels: document, text node and
within a node.
1. An annotation is mapped to its original document
using W3C’s URI (Uniform Resource Identifier)
addressing
8
. For example:
2. Within an XML document, text nodes are ad-
dressed using XPath. For example, the following
XPath expressions map to the title and the abstract,
respectively, of the MEDLINE citation whose
PMID is 12345, in a given MEDLINE XML file:
3. Using the monad concept, mapping to a location
within a node is trivial. It is done by simply list-
ing the monads of an annotation. For example,
given the context in Fig. 2, some of the annota-
tions in Fig. 1 may look like the following:
Annotation inclusion. An annotation file can
use annotation objects defined in other annotation
files. Suppose the
entity objects in Fig. 1 are defined
in a file named entities.xml. Then other files can
include entities.xml and reference the entities by
their names, prefixed with a unique identifier string
assigned to entities.xml. For example:
Token
: …protein kinase C (PKC) alpha, but not beta, isoform inhibited Akt phosphorylation…
Monad
:
…
12 13 14 15 16 17 18 19 20
21 22 23
…
Figure 2: Monads tokenized at white space.
#include “entities.xml” prefix “a”
…
event[a] = { actor1 = a:entity[3],
action = {23},
actor2 = null};
event[b] = { actor1 = a:entity[1],
action = {21},
actor2 = event[a]
}
;
//MedlineCitation[PMID=12345]//ArticleTitle
//MedlineCitation[PMID=12345]//AbstractText
context.file = <file address>;
context.node = <node address>;
context.delimiters = “ ”;
context.includeDelimiters = no;
entity[1] = {12, 13, 14, 16, 20};
entity[2] = {12, 13, 14, 19, 20};
entity[3] = {22};
event[a] = { actor1 = entity[3],
action = {23},
actor2 = null};
event[b] = { actor1 = entity[1],
action = {21},
actor2 = event[a]};
http://www.iastate.edu/~berleant/med35.xml
ftp://www.pub.iastate.edu/users/index.htm
file://c:\My Documents\manuscript\SOOML.txt
ICEIS 2005 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
384

4 FULFILLING THE
REQUIREMENTS
The four mechanisms work closely together to meet
all of the text-mining annotation requirements.
Expressiveness: The requirement of expressive
power has two aspects–the need to express complex
hierarchical structures and the need to express arbi-
trarily distributed surface strings. For the former,
the object-oriented mechanism enables modeling of
complex hierarchical structures, similar to the use of
object-oriented programming languages in modeling
software application environments. For the latter, the
monad-based mapping mechanism enables listing
the tokens anywhere in a document.
Resolution of annotation mapping: Monad-
based mapping also enables annotation mapping at
various resolutions. Monads are the atomic units in
SOOML. Their sizes, hence the mapping resolu-
tions, are determined by the set of delimiting charac-
ters. For example, “insulin-induced” can be treated
as a single monad (an adjective) in linguistic part-of-
speech tagging, so it need not be tokenized at the
character “-.” However, this resolution is not
enough for protein name recognition. Therefore, it
should be split into two monads (“-” being a delim-
iter). The finest resolution is single character map-
ping (using “null” delimiting character).
Reusability: The inclusion mechanism enables
annotation reuse, facilitating modular design of
complex text-mining systems in accordance with
software engineering principles. It is not necessary
to design a single powerful super-module to extract
the information all at once. Specialized modules can
target particular aspects of a complicated task and
create annotations on top of each other. The standoff
mechanism leaves the original documents un-
changed, thereby avoiding interference among dif-
ferent modules and/or applications.
Independence of availability: The standoff
mechanism separates annotations from the original
documents, and the monad-based mapping mecha-
nism avoids copying any contents from the original
text. Therefore, the availability of the annotations is
independent of the originals.
Flexibility: SOOML’s mechanisms enable anno-
tating files of various formats in a consistent way
(“one shoe fits all”). First, the standoff mechanism
separates annotations from the original documents;
therefore, the formats and the organization of the
annotations are not restricted by those of the original
documents. In contrast, in-line markup methods
have to follow the formats of the original texts. Sec-
ond, although SOOML’s mapping mechanisms (es-
8
http://www.w3.org/Addressing
pecially node-level mapping using XPath) are de-
signed for XML-based original documents, they can
be extended easily to any document with well-
defined fields or sections, because they are concep-
tually equivalent to the nodes in XML documents.
The worst case is for those files without any appar-
ent internal structures. SOOML can still treat such a
file as a single large text node, and monadize it from
the first token to the last.
Efficiency: The monad-based mapping mecha-
nism is space efficient. Instead of copying the con-
tent from the original documents, it uses monads
(equivalent to pointers) pointing to the sources. For
example, it takes only five integers to mark up
en-
tity[1]
in Fig. 1, while XPointer needs over 200 char-
acters. This makes SOOML an ideal format for an-
notation storage and exchange, as well as for serving
as an intermediate data-flow format among the mod-
ules/applications.
The monad-based mapping mechanism also
greatly reduces the complexity of annotation proc-
essing. First, gapped and overlapped annotations are
handled in exactly the same way as continuous and
non-overlapped ones. Second, monad-based map-
ping does not create any ambiguities, which are in-
evitable in string matching-based processing.
Extensibility: Because it is object oriented,
SOOML can be integrated readily with other ontolo-
gies. Ontologies typically already have well-defined
hierarchical structures. All we need to do is define
the main ontology entries as subclasses of the
anno-
tation
class (or one of its subclasses). The rest of the
ontology is automatically included in the hierarchy.
In conclusion, we presented here the design of a
standoff object-oriented markup language (SOOML),
which provides an expressive, efficient, flexible and
extensible framework for text annotation in bioin-
formatics – as well as other similar applications.
REFERENCES
Bird, S. and Liberman, M. (1999) A Formal Framework
for Linguistic Annotation. Technical Report MS-CIS-
99-01, Department of Computer and Information Sci-
ence, University of Pennsylvania.
Doedens, C.-J. (1994) in Text Databases. One Database
Model and Several Retrieval Languages. Amsterdam
and Atlanta, GA.
Hucka, M., et al. (2003) The Systems Biology Markup
Language (SBML): A Medium for Representation and
Exchange of Biochemical Network Models. Bioinfor-
matics 19: 524-531.
Kim, J.D., Ohta, T., Tateisi, Y., and Tsujii, J. (2003)
GENIA Corpus - A Semantically Annotated Corpus
for Bio-textmining. Bioinformatics 19: i180-i182.
DESIGN OF A STANDOFF OBJECT-ORIENTED MARKUP LANGUAGE (SOOML) FOR ANNOTATING
BIOMEDICAL LITERATURE
385
