
AUTOMATIC IDENTIFICATION OF NEGATED CONCEPTS IN
NARRATIVE CLINICAL REPORTS
Lior Rokach
Department of Information Systems Engineering, Ben-Gurion University of the Negev, Beer-Sheva, Israel
Roni Romano, Oded Maimon
Department of Industrial Engineering, Tel Aviv University, Israel
Keywords: Medical Informatics, Text Classification, Machine Learning, Information Retrieval.
Abstract: Substantial medical data such as discharge summaries and operative reports are stored in textual form.
Databases containing free-text clinical narratives reports often need to be retrieved to find relevant
information for clinical and research purposes. Terms that appear in these documents tend to appear in
different contexts. The context of negation, a negative finding, is of special importance, since many of the
most frequently described findings are those denied by the patient or subsequently “ruled out.” Hence, when
searching free-text narratives for patients with a certain medical condition, if negation is not taken into
account, many of the documents retrieved will be irrelevant. In this paper we examine the applicability of
machine learning methods for automatic identification of negative context patterns in clinical narratives
reports. We suggest two new simple algorithms and compare their performance with standard machine
learning techniques such as neural networks and decision trees. The proposed algorithms significantly
improve the performance of information retrieval done on medical narratives.
1 INTRODUCTION
Medical narratives present some unique problems.
When a physician writes an encounter note, a highly
telegraphic form of language may be used. There are
often very few (if any) grammatically correct
sentences, and acronyms and abbreviations are
frequently used. Very few of these abbreviations and
acronyms can be found in a dictionary and they are
highly idiosyncratic to the domain and local
practice. Often misspellings, errors in phraseology,
and transcription errors are found in dictated reports.
Researchers in medical informatics suggested
methods for automatically extracting information
contained in narrative reports for decision support
(Fiszman et al., 2000), guideline implementation
(Fiszman and Haug, 2000), and detection and
management of epidemics (Hripcsak et al., 1999).
Nevertheless most of the researches have
concentrates on methods for improving information
retrieval from narrative reports (see for instance,
Hersh and Hickam, 1995; Nadkarni, 2000; Rokach
et al., 2004). A search for patients with a specific
symptom or set of findings might result in numerous
records retrieved. The mere presence of a search
term in the text, however, does not imply that
records retrieved are indeed relevant to the query.
Depending upon the various contexts that a term
might have, only a small portion of the retrieved
records may actually be relevant.
A number of investigators have tried to cope
with the problem of a negative context. Aronow et
al. (1999) developed the NegExpander which uses
syntactic methods to identify negation in order to
classify radiology (mammography) reports. While
NegExpander is simple in that it recognizes a limited
set of negating phrases, it does carry out expansion
of concept-lists negated by a single negating phrase.
Friedman et al. (1994) developed the MedLEE
that performs sophisticated concept extraction in the
radiology domain. The MedLEE system combines a
syntactic parser with a semantic model of the
domain. MedLEE recognizes negatives which are
followed by words or phrases that represent specific
semantic classes such as degree of certainty,
temporal change or a clinical finding. It also
identifies patterns where only the following verb is
negated and not a semantic class (i.e. “X is not
increased”).
257
Rokach L., Romano R. and Maimon O. (2006).
AUTOMATIC IDENTIFICATION OF NEGATED CONCEPTS IN NARRATIVE CLINICAL REPORTS.
In Proceedings of the Eighth International Conference on Enterprise Information Systems - AIDSS, pages 257-262
DOI: 10.5220/0002497702570262
Copyright
c
SciTePress

Mutalik et al. (2001) used a lexical scanner with
regular expressions and a parser that uses a restricted
context-free grammar to identify pertinent negatives
in discharge summaries and surgical notes. Their
system first identifies propositions or concepts and
then determines whether the concepts are negated.
The set of regular expressions is predefined by IT
professional based on input obtained from medically
trained observers.
Chapman et al. (2001) developed a simple
regular expression algorithm called NegEx that
implements several phrases indicating negation,
filters out sentences containing phrases that falsely
appear to be negation phrases, and limits the scope
of the negation phrases. Their algorithm uses a
predefined set of pseudo negation phrases, a set of
negation phrases, and two simple regular
expressions.
There is no research that tries to learn the
negation patterns automatically and then uses the
discovered patterns to classify medical concepts that
appears in unseen texts.
Physicians are trained to convey the salient
features of a case concisely and unambiguously as
the cost of miscommunication can be very high.
Thus it is assumed that negations in dictated medical
narrative are unlikely to cross sentence boundaries,
and are also likely to be simple in structure (Mutalik
et al., 2001). Based on the above assumptions the
purpose of this work is to develop a methodology for
learning negative context patterns in medical
narratives and measure the effect of context
identification on the performance of medical
information retrieval.
2 MACHINE LEARNING
FRAMEWORK
The proposed process begins by performing several
preprocessing steps. First all medical documents
were parsed. Then all known medical terms are
tagged using a tagging procedure presented in
(Rokach et al., 2004). Finally each text was broken
into sentences using a sentence boundary identifier
as suggested in (Averbuch et al., 2003).
Physician reviewed each document and labelled
each medical term indicating whether it appear in
positive or negative context. Note that there might
be several medical terms in a single sentence not
necessarily with the same label. Consider for in-
stance the compound sentence "The patient states
she had fever, but denies any chest pain or shortness
of breath" In this case "chest pain" and "shortness of
breath" are negative while "fever" is positive.
The resulting labelled dataset was divided into 2
sets: the training set which contained the cases of
two-thirds of the documents. The remaining cases
are used as a test set.
The training set serves as the input to the
learning algorithm. The output of the learning
algorithm is a classifier. Given a tagged sentence
and a pointer to a tagged term, the classifier
classifies the indicated tagged term to either negative
or positive context.
In this section we present several learning
algorithms that can be used to classify a given
medical term into positive or negative context. We
begin by accommodating standard text classification
algorithms to the problem examined here. Then we
propose two new algorithms developed specifically
for this problem.
2.1 Standard Learning Algorithms
The most straightforward approach is to use existing
supervised learning algorithms. In fact the problem
presented here is a specific case of text classification
task. A detailed overview of text classification can
be found in Sebastiani (2002).
The main problem, in comparison to
conventional classification tasks, is the additional
degree of freedom that results from the need to
extract a suitable feature set for the classification
task. Typically, each word is considered as a
separate feature with either a Boolean value
indicating whether the word occurs or does not
occur in the document (set-of-words representation)
or a numeric value that indicates the frequency (bag-
of-words representation).
In this research we are using the bag-of-words
representation. Nevertheless instead of using a single
bag-of-words representation for the entire sentence,
we are using two bags: one for the words that
precede the targeted medical term and one for the
words that follow it. This split may help to resolve
some of the identification problems that arise in
compound sentences that include both positive and
negative in the same sentence. Recall the example
"The patient states she had fever, but denies any
chest pain or shortness of breath". In this case the
appearance of the verb "denies" after the term
"fever" indicates that the term "fever" is left in
positive context.
In the experimental study presented bellow we
examine the following induction algorithms:
Decision Tree using the C4.5 algorithm (Quinlan,
1993), Naïve Bayes (Duda and Hart, 1973), Support
ICEIS 2006 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
258

Vector Machines using the improved Platt's SMO
Algorithm (Keerthi et al.), Neural Networks and
Logistic Regression with a ridge estimator (Cessie
and van Houwelingen, 1997)
2.2 Profile Based Learning
Algorithm
We now suggest a simple algorithm that uses
information theory to find the negative context
profile. The profile consists of a list of indicating
terms. For instance the profile can be the
L={"negative for", "denies"}. This profile is then
used to classify new instances.
All words or phrases that appear in the same
sentence as the targeted term are put on a list and
statistics are generated regarding their appearances
in negative and positive contexts. This list is then
filtered using a threshold parameter, to eliminate
rare words or phrases. Moreover all tagged terms are
also removed. The next step is calculating the
information gain (IG) for each term in each context.
Equation 1 shows how IG is calculated for training
set T:
(, ) () ( | )IG T term H T H T term=−
(1)
where H(T) is the entropy and H(T|term) is
conditional entropy given the term:
(2)
where:
()
P
term
- the proportion of cases of T in which the
term appears.
()Pterm
- the proportion of cases of T in which the
term does not appear.
P
i
- the proportion of cases of T in which the context
was i (positive or negative).
P
i
(term) - the proportion of cases of T in which the
context was i and the term appears.
The last step of the algorithm is to remove from each
context profile, terms whose IG is below a certain
threshold.
2.3 Regular Expression Learning
The basis for discovering a regular expression is a
method that compares two texts with the same
context and incorporates the same concept types (i.e.
diagnosis, medication, procedure, etc.). By
employing the Longest Common Subsequence
algorithm (Myers, 1986) on each part of the sentence
(before the targeted term and after the targeted term)
a regular expression that fits these two sentences is
created. For instance let's look on the following two
sentences:
The patient was therefore admitted to the
hospital and started on Vancomycin as
treatments for endocarditis.
The patient was ruled in for myocardial
infarction and started Heparin for unstable
angina.
In this case the expert can point on the
"Vancomycin" and "heparin" as positive context of
medication. Thus we can execute the Longest
Common Subsequence algorithm on the two pairs of
strings (before and after the targeted term) presented
in Table 1.
Table 1: Longest Common Substring Searching.
Sentence 1 Sentence 2
The patient was
therefore admitted to
the hospital and
started on
The patient was ruled in
for <DIAGNOSIS> and
started
as treatments for
<DIAGNOSIS>.
for
<DIAGNOSIS>.
As a result of running the Longest Common
Subsequence algorithm we can obtain the following
pattern. This pattern can now be used to classify
concept of type medication appearing in positive
contexts.
The patient was [^.]{0,40} and started
[^.]{0,3} <MEDICINE> [^.]{0,14} for
<DIAGNOSIS>
Obviously there are many patterns that can be
created (each pair of sentences with the same
concept type and context). Thus we need a criterion
to select the pattern that best differentiate the
negative context from the positive context. For this
purpose we validate the generalization of the pattern
of concept type by calculating the information gain.
Enumerating over all candidate patterns we select
the pattern with the highest information gain (we
denote it as best_pattern). Following that we
recursively look for a new regular pattern in each of
the two possible outcomes of best_pattern. Namely
we find a pattern for all cases that implement
best_pattern and a pattern for all cases that do not
implement best_pattern. The procedure is repeated
in a recursive manner until no improvement in
information gain can be obtained. This procedure
creates a decision-tree-like structure of patterns for
each concept type.
2
{,}
2
{,}
( ) ()* ()log()
()* ()log()
ii
i pos neg
ii
i pos neg
H T term P term P term P term
Pterm P term P term
∈
∈
=− −
∑
∑
AUTOMATIC IDENTIFICATION OF NEGATED CONCEPTS IN NARRATIVE CLINICAL REPORTS
259

3 EXPERIMENTAL STUDY
The potential of the proposed methods for use in real
word applications was studied. In this experimental
study we used 4129 fully de-identified discharge
summaries that were obtained from Mount Sinai
Hospital in New-York. The database was divided
into two groups using a 2:1 ratio. The training set
consisted of 2752 documents (two-thirds of the total)
and the test set contained 1377 documents.
A physician was asked to label the following
terms “Nausea”, “Abdominal Pain”, “Weight Loss”
and “Diabetes Mellitus” in the training set. In
addition, the following terms were labeled in the test
set: “Headache”, “Hypertension” and “Chills.”
This list of terms was chosen to represent
different aspects of medical queries: simple terms
(e.g., nausea), terms that contain more than one
word, very popular terms, and ones that are
measured with numerical values (e.g., 10 pound
weight loss). Note that we used different terms in the
training set and in the test set in order to best
measure the generalization capability of the learning
algorithm.
Each appearance of the above terms was labelled
as having either a positive or negative context.
Table 2 presents the distribution of the two
contexts in the training set. The distribution is
measured both in terms of documents and in terms
of appearances (i.e., a given term can appear more
than once in the same document).
3.1 Measures Examined
The first measure used is the well-known
misclassification rate, indicating the portion of terms
that were misclassified by the classifier that was
created by the examined algorithm.
Additionally because the identification of the
negated is mainly used for improving information
retrieval, we will also examine the well-known
performance measures precision (P) and recall (R).
The notion of "precision" and "recall" are widely
used in information retrieval (Van Rijsbergen, 1979)
and data mining. Statistics use complementary
measures known as "type-I error" and "type-II
error".
Precision measures how many cases classified
as "positive" context are indeed "positive". Recall
measures how many "positive" cases are correctly
classified. Usually there is a trade-off between the
precision and the recall. Trying to improve one
measure often results in a deterioration of the second
measure. Thus, it is useful to use their harmonic
mean known as F-Measure.
The retrieval part of the experiment was meant to
simulate queries made by physicians. All the
documents in the test set were scanned for the query
terms. In each document where query terms were
found, a context classification, either positive or
negative, was made for each appearance of the term.
The context was classified by searching all the terms
of the sentence where the query term was found and
comparing it to the negative context profile. If a
term was found in the negative context profile, that
appearance of the query term was marked as
negative. After classifying all appearances of the
query terms in a document, the document was
retrieved only if at least one appearance of the query
term was in a ‘positive’ context.
Additionally, we measured the performance of
context insensitive retrieval; namely, assuming that
the context is always positive. The last measurement
can be useful for determining the impact of context
in medical narratives.
3.2 Results
Table 3 presents the mean F-Measure and
misclassification rate (over all queries) obtained by
each method on all medical terms. The results
indicate that the proposed algorithms have obtained
the highest F-Measure and the lowest
misclassification rate. Both algorithms are located in
the Pareto-graph. Decision Trees and Support Vector
has achieved the second best result.
Table 2: Context Distribution in the Training Set.
Term Positive
context
(documents)
Positive
context
(appearances)
Negative
context
(documents)
Negative
context
(appearances)
Nausea 284 370 251 286
Abdominal
pain
210 284 82 91
Weight
loss
94 108 21 21
Diabetes
mellitus
605 970 535 620
ICEIS 2006 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
260
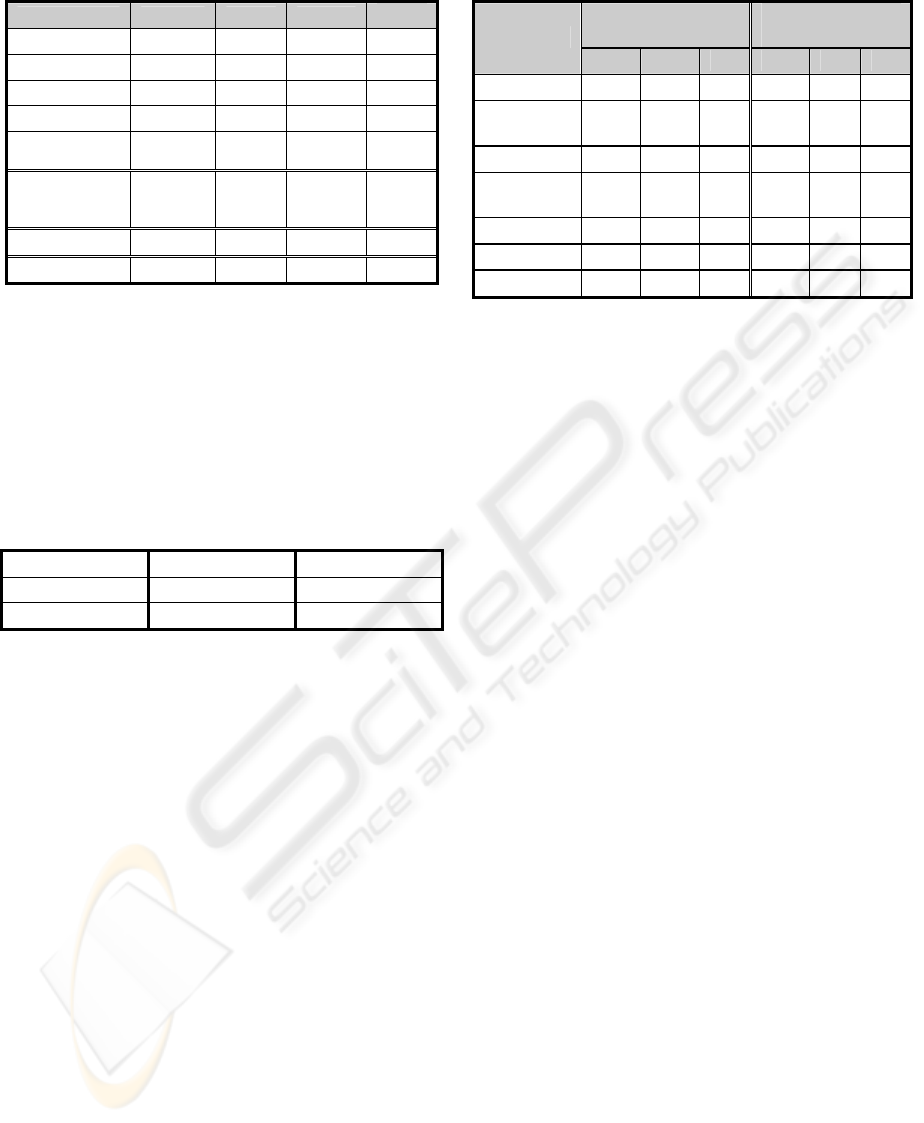
Table 3: Benchmark Results.
Method P R F
Error
Decision Tree 90% 92% 90.99%
10.40%
SVM 94% 88% 90.59%
10.40%
Naïve Bayes 82% 93% 87.15%
15.60%
Logistic Reg. 79% 86% 82.53% 21%
Neural
Network
63% 98% 76.46% 34%
Context
Insensitive
Retrieval
54% 100% 60.65% 42%
Profile Based 99% 95% 97.47% 2.80%
Regular Exp. 99% 97% 97.90% 2.30%
Table 4 presents the negative context profile
obtained by the Profile Based Learning Algorithm.
This profile contains only ten words/phrases. Most
of the entries in the table are related to the negative
context. It is interesting to note that the term "no"
and "not" are not included in this profile. This is
because their solely appearance is not a sufficient
indication for negation.
Table 4: Profile Content for Negative Context.
Any denies of systems
Change in had no was no
Changes negative for without
Table 5 presents the performance obtained by
Profile Based Learning Algorithm and by the best
standard algorithm as appeared in Table 3 (decision
tree) for each query used. The table indicates that the
proposed algorithm obtains better result in all
queries. Furthermore, the proposed algorithm has
relatively small variance. Table 5 also indicates that
the results obtained by the proposed algorithm for
the previously unseen terms (“Headache”,
“Hypertension” and “Chills”) and the remaining
terms (“Nausea”, “Abdominal Pain”, “Weight loss”
and “Diabetes Mellitus”) are similar.
The results of the decision tree classification
were compared to the ones obtained by the Profile
Based Learning Algorithm using McNemar’s test,
with continuity correction. The Chi squared obtained
was 11.172 with one degree of freedom. The two-
tailed P value was 0.0008. By conventional criteria,
this difference is considered to be statistically
significant.
Table 5: Performance by Term.
Decision Tree
Profile Based
Learning Algorithm
Query
P R F P R F
Nausea 96% 96% 96% 100% 98% 99%
Abdominal
Pain
96% 97% 96% 100% 96% 98%
Weight Loss 88% 100% 94% 100% 91% 95%
Diabetes
Mellitus
89% 92% 90% 98% 93% 95%
Headache 92% 95% 94% 100% 96% 98%
Hypertension 83% 94% 88% 100% 98% 99%
Chills 88% 98% 93% 97% 94% 96%
3.3 Error Analysis
Analyzing the reasons for False-Positive and False-
Negative results indicate that there are five main
categories of error:
Compound Sentence—Compound sentences are
composed of two or more independent clauses that
are joined by a coordinating conjunction or a
semicolon.
Reference to the Future — In this type of
sentence, the patient is given instructions on how to
react to a symptom he may develop, but currently
lacks. For example: “The patient was given clear
instructions to call for any worsening pain, fever,
chills, bleeding.” In this case the patient does not
suffer from fever, chills or bleeding and a query for
one of these symptoms will mistakenly retrieve the
document.
Negation indicating existence—Although the
meaning of a word might be negative, the context in
which it is written might indicate otherwise. For
example: “The patient could not tolerate the nausea
and vomiting associated with Carboplatin.”
Positive adjective—A sentence is written in a
negative form, but an adjective prior to one of the
medical term actually indicates its existence. For
example: “There were no fevers, headache or
dizziness at home and no diffuse abdominal pain,
fair appetite with significant weight loss.” The
adjectives “fair” and “significant” in the sentence
indicates that the following symptoms actually do
exist.
Wrong sentence boundaries—Sometimes the
boundary of a sentence is not identified correctly. In
this case, one sentence is broken into two, or two
sentences are considered as one.
Figure 1 presents the distribution of errors in the
test set for the Profile Based Learning Algorithm. It
AUTOMATIC IDENTIFICATION OF NEGATED CONCEPTS IN NARRATIVE CLINICAL REPORTS
261
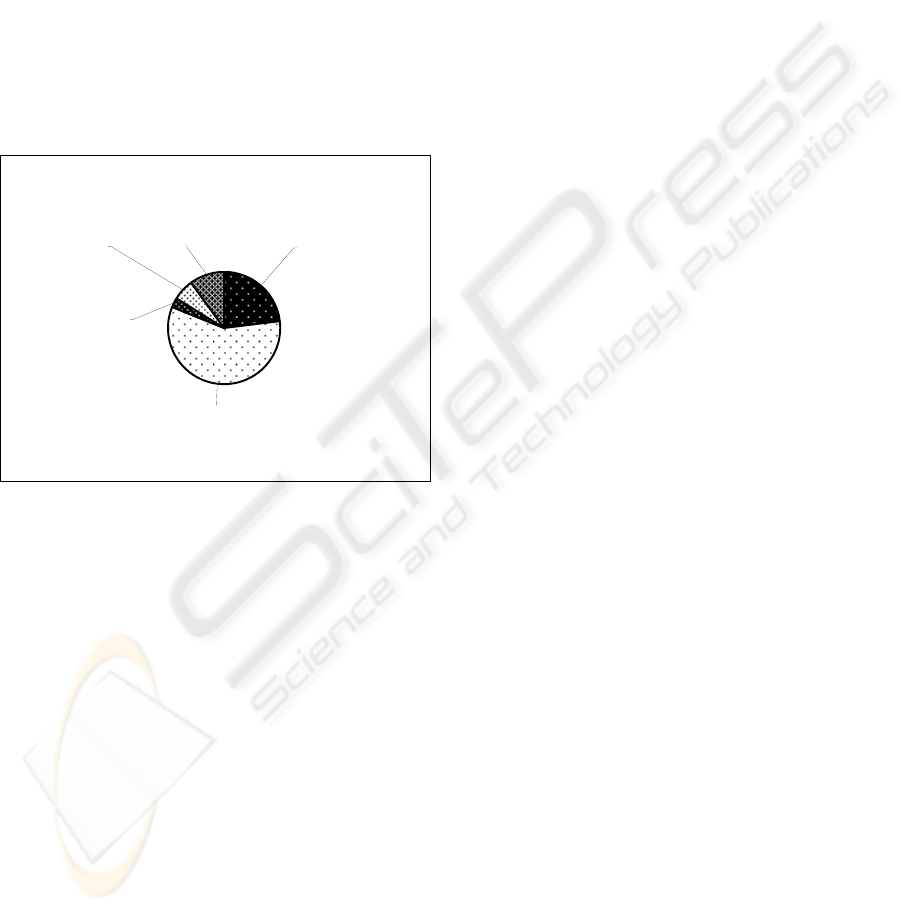
can be seen that the “compound sentence” is
responsible for most of the errors.
4 CONCLUSION
Two new algorithms for identifying context in free-
text medical narratives are presented. It has been
shown that the new algorithms are superior to
traditional text classification algorithms for common
medical terms such as: Nausea, Abdominal pain,
Weight loss etc. Furthers research can be made in
order to test the suggested algorithms for any
medical concept. The Profile Based Learning
Algorithm is also very simple but still outperforms
other more complicated methods.
Reference to
the future
10%
Wrong
sentence
boundaries
6%
Negation
indicating
existence
3%
Compound
sentence
58%
Positive
adjective
23%
Figure 1: Distribution of Errors for the Profile Based
Learning Algorithm.
REFERENCES
Aronow D, Feng F, Croft WB. Ad Hoc Classification of
Radiology Reports. Journal of the American Medical
Informatics Association 1999; 6(5): 393-411.
Averbuch M, Karson T, Ben-Ami B, Maimon O. and
Rokach L., Context-Sensitive Medical Information
Retrieval, MEDINFO-2004, San Francisco, CA,
September 2004, IOS Press, pp. 282-286.
Cessie S. and van Houwelingen, J.C. , Ridge Estimators in
Logistic Regression. Applied Statistics 1997: 41 (1):
191-201.
Chapman W.W., Bridewell W., Hanbury P, Cooper GF,
Buchanann BG. A Simple Algorithm for Identifying
Negated Findings and Diseases in Discharge
Summaries. J. Biomedical Info. 2001: 34: 301–310.
Duda R. and Hart P., Pattern Classification and Scene
Analysis. Wiley, New York, 1973.
Fiszman M., Chapman W.W., Aronsky D., Evans RS,
Haug PJ., Automatic detection of acute bacterial
pneumonia from chest X-ray reports. J Am Med
Inform Assoc 2000; 7:593–604.
Fiszman M., Haug P.J., Using medical language
processing to support real-time evaluation of
pneumonia guidelines. Proc AMIA Symp 2000; 235–
239.
Friedman C., Alderson P, Austin J, Cimino J, Johnson S.
A General Natural-Language Text Processor for
Clinical Radiology. Journal of the American Medical
Informatics Association 1994; 1(2): 161-74.
Hersh WR, Hickam DH. Information retrieval in
medicine: the SAPHIRE experience. J. of the Am
Society of Information Science 1995: 46:743-7.
Hripcsak G, Knirsch CA, Jain NL, Stazesky RC, Pablos-
mendez A, Fulmer T. A health information network
for managing innercity tuberculosis: bridging clinical
care, public health, and home care. Comput Biomed
Res 1999; 32:67–76.
Keerthi S.S., Shevade S.K., Bhattacharyya C., Murth
K.R.K., Improvements to Platt's SMO Algorithm for
SVM Classifier Design. Neural Computation 2001:
13(3):637-649.
Lindbergh D.A.B., Humphreys B.L., The Unified Medical
Language System. In: van Bemmel JH and McCray
AT, eds. 1993 Yearbook of Medical Informatics.
IMIA, the Netherlands, 1993; pp. 41-51.
Mutalik P.G., Deshpande A., Nadkarni PM. Use of
general-purpose negation detection to augment
concept indexing of medical documents: a quantitative
study using the UMLS. J Am Med Inform Assoc
2001: 8(6): 598-609.
Myers E., An O(ND) difference algorithm and its
variations, Algorithmica Vol. 1 No. 2, 1986, p 251.
Nadkarni P., Information retrieval in medicine: overview
and applications. J. Postgraduate Med. 2000: 46 (2).
Pratt A.W. Medicine, computers, and linguistics.
Advanced Biomedical Engineering 1973: 3:97-140.
Quinlan, J. R. C4.5: Programs for Machine Learning.
Morgan Kaufmann, 1993.
Rokach L., Averbuch M., Maimon O., Information
Retrieval System for Medical Narrative Reports,
Lecture Notes in Artificial intelligence 3055, pp. 217-
228 Springer-Verlag, 2004.
Sebastiani F., Machine learning in automated text
categorization. ACM Comp. Surv., 34(1):1-47, 2002.
Van Rijsbergen, CJ.. Information Retrieval. 2nd edition,
London, Butterworths, 1979.
ICEIS 2006 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
262
