
AUTOMATED TUMOR SEGMENTATION USING LEVEL SET
METHODS
Stephane Lebonvallet, Sonia Khatchadourian and Su Ruan
CReSTIC,IUT de TROYES, 9 rue du Quebec,1026 TROYES CEDEX
Keywords:
Image, Partial differential equations, Level Sets, Segmentation.
Abstract:
In the framework of detection, diagnostic and treatment planning of the tumours, the Positron Emission To-
mography (PET) and Magnetic Resonance Imaging (MRI) have become the most efficient techniques for
body and brain examination. Radiologists take usually several hours to segment manually the region of in-
terest (ROI) on images to obtain some information about patient pathology. It is very time consuming. The
aim of our study is to propose an automatic solution to this problem to help the radiologist’s work. This paper
presents an approach of tumour segmentation based on a fast level set method. The results obtained by the
proposed method dealing with both PET and MRI images are encouraging.
1 INTRODUCTION
PET (Positron Emission Tomography) and MRI
(Magnetic Resonance Imaging) scans are both stan-
dard imaging tools that physicians use to pinpoint dis-
ease states in the body and the brain. A PET scan
demonstrates the biological function of the body be-
fore anatomical changes take place, while the MRI
scan provides usually information about the brain tu-
mor. Accurate and automatic tumor segmentation is
a very important issue in many applications of medi-
cal image system for quantitative studies. As known,
manual tracing by an expert of a tumor in 3D is not
only exceedingly time consuming, but also exhaust-
ing for experts leading to human errors. Therefore, an
automatic segmentation is necessary.
Many segmentation methods have already been
used in medical imaging field. A simple method is
the thresholding. (Malyapa et al., 2002) used a binary
threshold to segment tumorous areas in the pelvis re-
gion. This method allows the user to obtain results
very quickly but it is not very accurate. The seeded
region growing is one of segmentation methods. It
consists of choosing seeds, and growing them accord-
ing to a criteria. One solution adopted by (Adams
et al., 1994) is to choose the seeds as the local minima
and maxima and then to grow them until the regions
touch themselves. But when one need to delineate
just one region, one cannot venture to fill the entire
image. That is why (Zoroofi et al., 2004) have cho-
sen the seeds upon the histogram and then let grow
until the criteria was no longer verified. This work
has been used to the segmentation of a non-necrotic
femoral head. Mathematical morphology segmenta-
tion based on watersheds (Roerdink et al., 2001)is
proposed in (Mancas et al., 2004)to segment PET
/CT images. The principle of the watershed tech-
nique is to transform the gradient of a grayscale im-
age into a topographic surface. An iterative water-
sheds is used in (Mancas et al., 2004). A very com-
mon way to segment medical images is to use an at-
las of the humain body as a priori knowledge. (Bon-
diau et al., 2005), (Erhardt et al., 2001) and (Lorenzo
et al., 2004) have used such approach respectively on
the brain, the hip and the heart atlases. The statisti-
cal methods have been also studied to deal with the
medical images, because of noise presenting in im-
ages. (Ruan et al., 2000) propose a statistical segmen-
tation based on Random Markov Fields to segment
brain RM images. The fuzzy segmentation offers an
interesting point of view to take into account ambigu-
ities of objets to segment. As shown in (Dou et al.,
2004), membership functions are modelled to express
the fuzzy signals of the brain tumor observed in differ-
128
Lebonvallet S., Khatchadourian S. and Ruan S. (2007).
AUTOMATED TUMOR SEGMENTATION USING LEVEL SET METHODS.
In Proceedings of the Second International Conference on Computer Vision Theor y and Applications, pages 128-133
DOI: 10.5220/0002068301280133
Copyright
c
SciTePress

ent types of images. Among various image segmenta-
tion techniques, active contour model [2] has emerged
as a powerful tool for semi-automatic object segmen-
tation. The basic idea is to evolve a curve, subject
to constraints from a given image,for detecting inter-
esting objects in that image. It consists in the reso-
lution of systems of partial differential equations for
which interface propagation phenomenon has to be
described. The active contour models are often imple-
mented based on level set method ((Sethian, 1999)),
which is a powerful tool to capture deforming shape.
But it has the disadvantage of a heavy computation re-
quirement even using the narrowband evolution. The
fast marching method is proposed for monotonically
advanced fronts ((Sethian, 1999)), and is extremely
faster than level set evolution. Generally, there are
three key problems needed to be solved to implement
the curve evolution methods. The first one is the ini-
tialization of the seed points. The second one is the
formulation of the speed function. And the last one is
the determination of the stopping criterion.
The level set methods have been widely applied
in medical imagery (Suri et al., 2002) in different do-
mains : the brain (Xie et al., 2005), the bone (Morigi
et al., 2004), the vascular trees (Farag et al., 2004)
and so on... The most common way to initialize the
level set is the manual selection of a ROI which seems
to be relevant ((Xu et al., 2000), (Farag et al., 2004)
and (Xie et al., 2005)). Sometimes a simple mouse
click combined with a fast marching approach (Fan, )
is used. In this case the final contour determined by
the fast marching step is the initial front of the level
set. Those methods are semi-automated while we are
focused on the automated methods. (Morigi et al.,
2004) proposed an automated method but the imag-
ing system is not the same as the subject of our study.
Our work consists of detecting tumors from the
whole body image volume acquired by a PET/CT de-
vice. We have no a priori knowledge on the loca-
tion of the tumor zone to detect. A contour evolution
model using a level set method with an initialization
based on thresholding is proposed in this paper.
The paper is organized as the following. Firstly,
an overview of our study is described. Secondly, the
principle of the level set method and its implementa-
tion will be exposed. The different steps of our ap-
proach and the associated results are then presented.
Finally we will conclude and give some perspectives.
2 OVERVIEW OF THE STUDY
2.1 Segmentation Framework
Our aim is at detecting the tumorous areas in the body
and in the brain from PET images and MRI images.
Any a priori knowledge about locations of the tumors
are taken into account. As the PET images are usually
noisy and bad contrasted, the methods based on the
image intensity or gradient are not efficient in these
cases. The statistical methods cannot be neither used
efficiently due to the small size of the tumor : they
are too small to get statistical properties comparing
with all images. The solution of the evolving contours
is interesting in this case because they can grow to
the expected size of the tumorous areas with help of
geometrical and intrinsic properties.
For segmenting the 3D images, we complete it
through a 2D slice-by-slice process. The proposed
framework consists of 3 steps: seed detection giving
a set of seeds which are susceptible belonging to the
tumor; seed selection allowing to obtain one seed con-
sidered as the initial tumor contour; contour evolution
according to an active contour model.
The seed detection consists of finding ROIs us-
ing intensity information. The areas of high glucose
activity lead to high gray levels observed in PET im-
ages. A thresholding of images can be carried out to
obtain the ROIs. The problem is how to choose the
threshold. As known, the histogram can give the in-
formation about the distribution of grey levels. The
maximum of the histogram is firstly found, which rep-
resents body tissus. Supposing that the number of pix-
els belonging to tumor regions has less than that of the
pique of histogram. The threshold is then defined as
the gray level on which the number of pixels equals to
the maximum multiplied by a proportionality factor α
which is given by experiences.
After the thresholding of images, several seeds are
obtained in which some of them do not belong to the
tumor. The big regions representing some anatomi-
cal regions which give high intensity, and very small
regions due to noise, are detected as seeds. The big
regions can be easily moved out from the seeds. A di-
latation, morphological mathematics operator, is car-
ried out to eliminate the small seeds. This seed se-
lection step allows us to delete aberrant seeds and to
keep that of tumorous areas. From the obtained initial
contours (seeds), a level set method is used to grow
them to find the tumor contours. In the next section,
this method is presented in details.
AUTOMATED TUMOR SEGMENTATION USING LEVEL SET METHODS
129

3 CONTOUR EVOLUTION
BASED ON LEVEL SET
METHOD
The level set method has been introduced by (Os-
her et al., 1988) in order to solve the partial dif-
ferential equations. It refers to the theory of the
curve evolution. The algorithm proposed by Sethia
((Sethian, 1999) has been widely applied to many do-
mains ((Fan, ), (Xu et al., 2000))
3.1 Theory
Considering a curve represented by a level set func-
tion Φ, which is defined as a distance function :
For a point p, if Φ > 0, p is outside the contour,
if Φ = 0, p is on the contour,
if Φ < 0, p is inside the contour.
From a geometric point of view, the evolution of a
contour can be described as follows :
∂x
∂t
= VN (1)
with x a point of the contour, V the speed function, N
the normal vector of the curve at x. The evolution of
the curve depends on the normal vector N of the curve
and the curvature K at each point of the curve, with :
N =
∇Φ
|∇Φ|
K = ∇
∇Φ
|∇Φ|
=
Φ
xx
Φ
2
y
− 2Φ
x
Φ
y
Φ
xy
+ Φ
yy
Φ
2
x
(Φ
2
x
+ Φ
2
y
)
3/2
To describe the evolution of the curve we need to
initialize it. The initial curve is defined as zero level
set :
Φ((x(t)),t = 0) = 0 (2)
To associate the zero level set to the evolving
curve at each time and to derive the motion equation
for this level set function, the zero level set has to te
be re-initialized at each time step :
Φ((x(t)),t) = 0 (3)
After derivation of the equation 3 :
Φ
t
+V∇Φ((x(t)),t) = 0 (4)
where V = x
′
(t) · N is defined as a speed function.
This speed function is the key of the implementation
of the level set method.
3.2 Speed Function
The speed function V depends on :
• local properties given by local geometrical infor-
mation (curvature, normal of the curve),
• global properties depending on the form and the
position of the front,
• independent properties defined as a fluid velocity
that transport passively the front.
Based on these properties, the speed function can
be expressed as follows :
V = V
prop
+V
curv
+V
adv
(5)
with V
prop
= V
0
constant speed propagation
V
curv
= −εK curvature dependent speed
V
adv
= U(x,y,t) · N advection speed
Since the speed function decreases to zero at the
boundary of the area to segment, the components of
the speed function are proposed as follows:
ε = ε
c
V
pij
with ε
c
constant , (6)
V
pij
=
1
1+ Gij
with G
ij
the image gradient(7)
at pixel (i, j) ,
U
ij
= β∇V
pij
with β constant . (8)
ε
c
and beta are constant parameters to be defined
according to used images. For further details on the
implementation of the speed function, see (Sethian,
1999) and (Xu et al., 2000).The stopping criteria de-
pends on the speed function, therefore indirectly on
the intrinsic parameters of the images. The evolution
of contours stops when the speed function V = 0.
3.3 The Narrow Band
The problem of this method is that it takes a long time
to compute if the update of the level set function is
made on the entire image. The solution proposed by
(Chopp, 1993) is to compute the level set function in a
narrow band around the front. The level set function
is only updated when it reaches the boundary of the
narrow band. This narrow band approach can reduce
importantly the computing time. It has been used in
shape recognition by (Malladi et al., 1994) and an-
alyzed by (Adalsteinsson et al., 1995) with success.
Therefore, the narrow band is also adopted to the our
method in the process of evolution.
VISAPP 2007 - International Conference on Computer Vision Theory and Applications
130

4 EXPERIMENTAL RESULTS
4.1 Data
The data PET, we use for our experimentation, con-
sist of three PET image volumes corresponding to
three patients who reach some tumors. The size of
the images is of 144x144 pixels and their resolution
is above 7mm per pixel for both PET and Ct images.
One volume is composed of about 190 slices. Visu-
ally, the three tumors observed in images are well seg-
mented, confirmed by hospital experts. The MRI im-
ages are acquired on a 1.5T GE (General Electric Co.)
machine using an axial FLAIR. The image volume
consists of 512 (pixels) x 512(pixels) x 24 (slices)
with a voxel size of 0.470.47 5.5 mm3. Three vol-
umes of one patient acquied during a medical treat-
ment at three time points are used in our experience.
Six months separates each time point. The volume
variations can be then calculated from segmentation
results.
4.2 Choice of Parameters
Different values of the parameters have studied and
tested before validate these ones : α = 0, 0025 for
the seed detection parametrization, ε
c
= 0,05 and β =
0,005 for the level set parametrization. Those values
are chosen according to the images to be dealed with.
They can keep the same if the images to be treated are
acquired from the same imaging machine.
4.3 Segmentation
Ten PET slices of a patient who reaches a lung tumor
are presented here (figure 1). For giving a good vi-
sualization, the gray levels are inverted. The tumor
appears dark. The results obtained in different steps
are shown from the figure 2 to 4:
• seed detection,
• seed selection,
• tumor segmentation with the level set method.
The seed detection (figure 2) allows us to deter-
mine ROIs which could be contained by tumorous ar-
eas. It is achieved thanks to the gray level informa-
tion of the entire image volume. We know that higher
gray levels represent areas of higher glucose activity
and the tumors have abnormal glucose activity. But
as foreseen the ROIs obtained are not necessarily tu-
mors. The seed selection (figure 3) can help the deci-
sion of seed as explained previously.
The level set method is carried out image by image
for all the voulme from the initialization given by the
seed selection. We can see on figure 4 that the tumor
is well segmented. The same approche is performed
on MRI data. For example, for one patient, the varia-
tion is decreased about 15.42 percent six months after.
This measure provides a very important information
for the experts to evaluate the medical treatment.
5 CONCLUSION AND FUTURE
WORKS
This paper presents a work on the automatic segmen-
tation of tumorous areas for whole body and brain.
The tumors are well segmented even if it remains in
the results healthy regions. Two possibilities have
been evoked to solve this :
- Improving the seed detection by using a multi-scale
binarization method (see (Jolion, 1994) and (Trier
et al., 1995)) for example. Indeed the question of seed
detection has been briefly considered to test the level
set method.
- Implementing a very robust classification method.
A SVM classification has already evoked : previous
works on medical image classification, lead in the lab-
oratory, gave encouraging results (AitAouit, 2004).
If the initialization step is not robust enough, the clas-
sification step is necessary to select tumor contour.
Finally the level set should be implemented as a
real 3-dimensional method in order to consider the
whole 3D information. That allows to ameliorate the
performances of the coutour evolution.
REFERENCES
Adalsteinsson et al. (1995). A fast level set method for prop-
agating interfaces. Jour. of Comp. Phys., 118:269–
277.
Adams et al. (1994). Seeded region growing. IEEE
Transaction on Pattern Analysis Machine Intelligence,
16:641–647.
AitAouit (2004). Classification d’images par la mthodes des
support vector machines (svm): tude et applications.
Bondiau et al. (2005). Atlas-based automatic segmentation
of mr images: Validation study on brainstem in ra-
diotherapy context. Int. J. Radiation Oncology Biol.
Phys., 61:289–298.
Chopp, D. (1993). Computing minimal surfaces with level
set curvature flow. Jour. of Comp. Phys., 106:77–91.
Dou et al. (2004). Automatic brain tumor extraction using
fuzzy information fusion. Proc. SPIE, 4875:604–609.
Erhardt et al. (2001). Atlas-based segmentation of struc-
tures to support virtual planning of hip operations. In-
ternational Journal of Medical Informatics, 64:439–
447.
AUTOMATED TUMOR SEGMENTATION USING LEVEL SET METHODS
131

Fan, D. www.cs.wisc.edu/˜fan/levelset/.
Farag et al. (2004). 3d volume segmentation of mra data sets
using level sets. Academic Radiology, 11:419–435.
Jolion, J.-M. (1994). Analyse multirsolution du contraste
dans les images numriques. Traitement du Signal,
11:245–255.
Lorenzo et al. (2004). Segmentation of 4d cardiac mr im-
ages using a probabilistic atlas and the em algorithm.
Medical Image Analysis, 8:255–265.
Malladi et al. (1994). Evolutionnary fronts for topology-
independent shape modeling and recovery. Proceed-
ings of Third European Conference on Computer Vi-
sion, 800:3–13.
Malyapa et al. (2002). Physiologic fdg-pet three-
dimensional brachytherapy treatment planning for
cervical cancer. Int. J. Radiation Oncology Biol.
Phys., 54:1140–1146.
Mancas et al. (2004). Towards an automatic tumor segmen-
tation using iterative watersheds. Proc. of the Medical
Imaging Conference of the International Society for
Optical Imaging (SPIE Medical Imaging).
Morigi et al. (2004). 3d long bone reconstruction based on
level sets. Computerized Medical Imaging and Graph-
ics, 28:377–390.
Osher et al. (1988). Fronts propagating with curvature-
dependent speed : algorithms based on hamilton-
jacobi formulations. J. Computational Physics,
79:12–49.
Roerdink et al. (2001). The watershed transform: Defi-
nitions, algorithms parallelization strategies. Funda-
menta Informaticae, pages 187–228.
Ruan et al. (2000). Brain tissue classification of magnetic
resonance images using partial volume modeling.
IEEE Transactions on Medical Imaging, 19:1179–
1187.
Sethian, J. (1999). evel Set Methods and Fast Marching
Methods. Cambridge Univerity Press.
Suri et al. (2002). Shape recovery algorithms using level
sets in 2-d/3-d medical imagery: A state-of-the-art re-
view. IEEE Transaction on Information Technology in
Biomedicine, 6:8–28.
Trier et al. (1995). Evaluation of binarization methods for
document images. IEEE Trans. Pattern Anal. Mach.
Intell., 17:312–315.
Xie et al. (2005). Semi-automated brain tumor and edema
segmentation using mri. European Journal of Radiol-
ogy.
Xu et al. (2000). Medical image segmentation using de-
formable models. In Fitzpatrick, J. and Sonka, M.,
editors, Handbook of Medical Imaging – Volume 2:
Medical Image Processing and Analysis, pages 129–
174. SPIE Pres.
Zoroofi et al. (2004). Automated segmentation of necrotic
femoral head from 3d mr data. Computerized Medical
Imaging and Garphics, 28:267–278.
VISAPP 2007 - International Conference on Computer Vision Theory and Applications
132
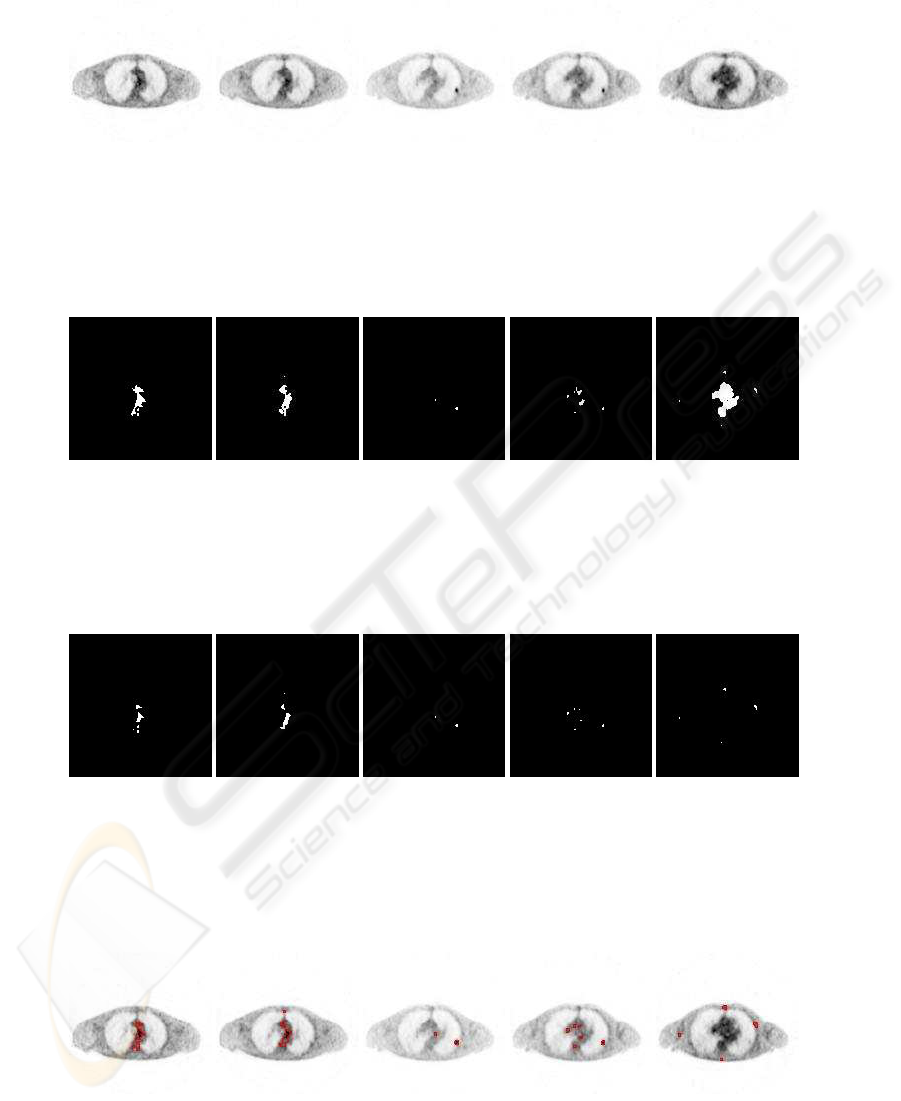
Figure 1: patient 3 stack sample illustrating a lung tumor.
Figure 2: Seed detection on this sample.
Figure 3: Seed treatment.
Figure 4: level set segmentation.
AUTOMATED TUMOR SEGMENTATION USING LEVEL SET METHODS
133
