
NAMED ENTITY RECOGNITION IN BIOMEDICAL LITERATURE
USING TWO-LAYER SUPPORT VECTOR MACHINES
Feng Liu, Yifei Chen
Computational Modeling Lab, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium
Bernard Manderick
Computational Modeling Lab, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium
Keywords:
Named entity recognition, gene/protein names identification, support vector machine, two-layer structure,
boundary check.
Abstract:
In this paper, we propose a named entity recognition system for biomedical literature using two-layer support
vector machines. In addition, we employ a post-processing module called a boundary check module to elim-
inate some boundary errors, which can lead to improved system performance. Our system doesn’t make use
of any external lexical resources and hence it is a fairly simple system. Furthermore, with carefully designed
features and introducing a second layer, our system can recognize named entities in biomedical literature with
fairly high accuracy, which can achieve the precision of 83.5%, recall of 80.8% and balanced F
β=1
score of
82.1%, an approximate state of the art performance for the moment.
1 INTRODUCTION
In recent years, there is a growing interest in genome
research. Hence the amount of biomedical literature
published daily is increasing exponentially. For ex-
ample, MEDLINE(MEDLINE, 1966), the primary re-
search database in biomedical domain, is an online
bibliographic source of citations and abstracts dating
from 1966 till present and now contains more than 14
million abstracts. Therefore, extracting information
from the biomedical literature manually is not real-
istic any more and an urgent demand for automated
information extraction system in biomedical domain
has emerged.
The focus of this paper is on named entity recog-
nition (NER) in biomedicine. Named entities in
biomedicine include names of genes, proteins, gene
products, organisms, etc. NER is an important first
step for information extraction in biomedical litera-
ture. Without precise recognition of these named en-
tities, it is impossible to extract the relations, infor-
mation and knowledge from the literature. However,
due to the complex biomedical nomenclature, NER in
biomedicine is often much more difficult than that in
newswire domain. For example, named entities can
be full names (such as “fatty acid synthase”) or ab-
breviations (such as “FAS”). Even worse, sometimes
abbreviations are not acronyms. Moreover, a given
named entity can have several aliases (e.g., a gene for
nerve growth factor receptor has at least 10 different
names). In order to handle these difficulties, many
different approaches have been applied in the past few
years, Support Vector Machines (SVMs) (Mitsumori
et al., 2005; Takeuchi and Collier, 2003), Hidden
Markov Models (HMMs) (Zhou et al., 2005; Morgan
et al., 2004) and rule-based systems (Tamames, 2005;
Tanabe and Wilbur, 2002). In general, there are 2 di-
visions for NER systems in biomedicine.
• Closed: The system is only trained on the training
data without any external lexicon.
• Open: The system can make use of external lexi-
cal resources.
Usually open systems are more complicated than
closed ones because open systems need incorporate
external lexicons. Moreover, it is not trivial to uti-
lize the lexicons properly, which heavily depends on
how to constitute the lexicons and how to design the
lexicon matching strategies. Some papers (Kinoshita
et al., 2005; Zhou et al., 2005) reported that adding the
lexicons could improve the performance while other
papers (Song et al., 2004; Tsuruoka and Tsujii, 2003)
39
Liu F., Chen Y. and Manderick B. (2007).
NAMED ENTITY RECOGNITION IN BIOMEDICAL LITERATURE USING TWO-LAYER SUPPORT VECTOR MACHINES.
In Proceedings of the Ninth International Conference on Enterprise Information Systems - AIDSS, pages 39-45
DOI: 10.5220/0002357300390045
Copyright
c
SciTePress

claimed that the performance even decreased after in-
corporating the lexicons.
Here we propose a closed system for recognizing
named entities from biomedical literature using two-
layer support vector machines without utilizing any
external lexicon. Our system is efficient due to its
simplicity and also effective because it can achieve
over 80% balanced F
β=1
score, an approximate state
of the art performance currently. We will discuss it in
detail in Section 2.
2 METHOD
2.1 Overview
Figure 1 shows our named entity recognition system.
We employ two SVMs to build a two-layer NER sys-
tem. The first layer is from text to entity, which takes
original text as input and recognizes named entities
from text. Then the second layer is from entity to en-
tity, which takes named entities from the first layer as
input, makes some corrections and outputs the final
recognition.
Figure 1: System Description.
2.2 Support Vector Machines
SVM (Vapnik, 1995) is a powerful machine learning
method, which was introduced by Vladimir Vapnik
and his colleagues in 1995. In SVM learning, the data
is mapped non-linearly from the original input space
X to a high-dimensional feature space F. Due to the
non-linear mapping to F, the complex non-linear de-
cision boundary in X becomes the simple linear deci-
sion boundary in F. Moreover, by introducing the ker-
nel function K, the mapping to F can stay implicit and
we can avoid working in the high-dimensional space
F; i.e., inner products in F can be directly calculated
by K taking vectors from the input space X and with-
out having to know the exact form of the mapping,
hence the implicit mapping and computational ben-
efit. Finally, the decision boundary is defined com-
pletely by inner products between vectors in F and
calculated through the kernel function K. Therefore,
in SVM learning, the most important design decision
is the choice of kernel function K.
In this paper we choose the toolbox LIBSVM
(Chang and Lin, 2001), a Jave/C++ library for SVM
learning, to implement our system. And we adopt the
following polynomial kernel as the kernel function:
X × X → R : K(
−→
x
i
,
−→
x
j
) = (
−→
x
i
·
−→
x
j
+ coef)
d
with coef = 0 and d = 2
2.3 Data Representation
The data used in our experiments is taken from the
training data of BioCreAtIvE II challenge (BioCre-
AtIvE, 2006), which consist of 15000 sentences de-
rived from MEDLINE abstracts that have been man-
ually annotated for names of genes/proteins and their
related entities.
The first pre-processing step is tokenization. Un-
like NER systems in newswire domain, here we can’t
remove punctuation to make processing straightfor-
ward because many gene/protein names contain hy-
phens, parentheses, brackets and other type of punc-
tuation. Hence we split the sentences based on spaces
and punctuation. For example, an sentence “the
M-phase-inducing Cdc2/Cdc13 cyclin-dependent ki-
nase” is tokenized into the following tokens: “the”,
“M”, “-”, “phase”, “-”, “inducing”, “Cdc2”, “/”,
“Cdc13”, “cyclin”, “-”, “dependent”, “kinase”.
Then the second pre-processing step is assigning
the class label for each token. We use the traditional
BIO representation as follows.
• B: current token is the beginning of a named entity
• I: current token is inside a named entity
• O: current token is outside a named entity
Consider that “Cdc2/Cdc13 cyclin-dependent ki-
nase” is a named entity. Therefore after pre-
processing, the above sentence becomes “O the”, “O
M”, “O -”, “Ophase”, “O -”, “O inducing”, “B Cdc2”,
“I /”, “I Cdc13”, “I cyclin”, “I -”, “I dependent”, “I ki-
nase”.
ICEIS 2007 - International Conference on Enterprise Information Systems
40

2.4 Feature Extraction
In the first layer, the following features are extracted
to represent each token.
• Token: token itself.
• Orthographic feature: Table 1 shows all the 22 or-
thographic features used in our system.
• POS: Part-of-speech of the current token. POS
can determine the syntactic functions of words
in text, which are very helpful for NER sys-
tems. However, POS taggers developed for
newswire domain don’t perform well when ap-
plied to biomedical literature. Hence we use the
MedPost tagger (Smith et al., 2004) in our system.
MedPost was trained on the MEDLINE corpus so
that it can achieve over 97% accuracy on MED-
LINE citations.
• Prefix: bi-, tri- and quad-prefix of the current to-
ken. For instance, the bi-, tri- and quad-prefix
of the token “transaminase” are “tr”, “tra” and
“tran”, respectively.
• Suffix: bi-, tri- and quad-suffix of the current to-
ken. Similarly, the bi-, tri- and quad-suffix of
“transaminase” are “se”, “ase” and “nase”, re-
spectively.
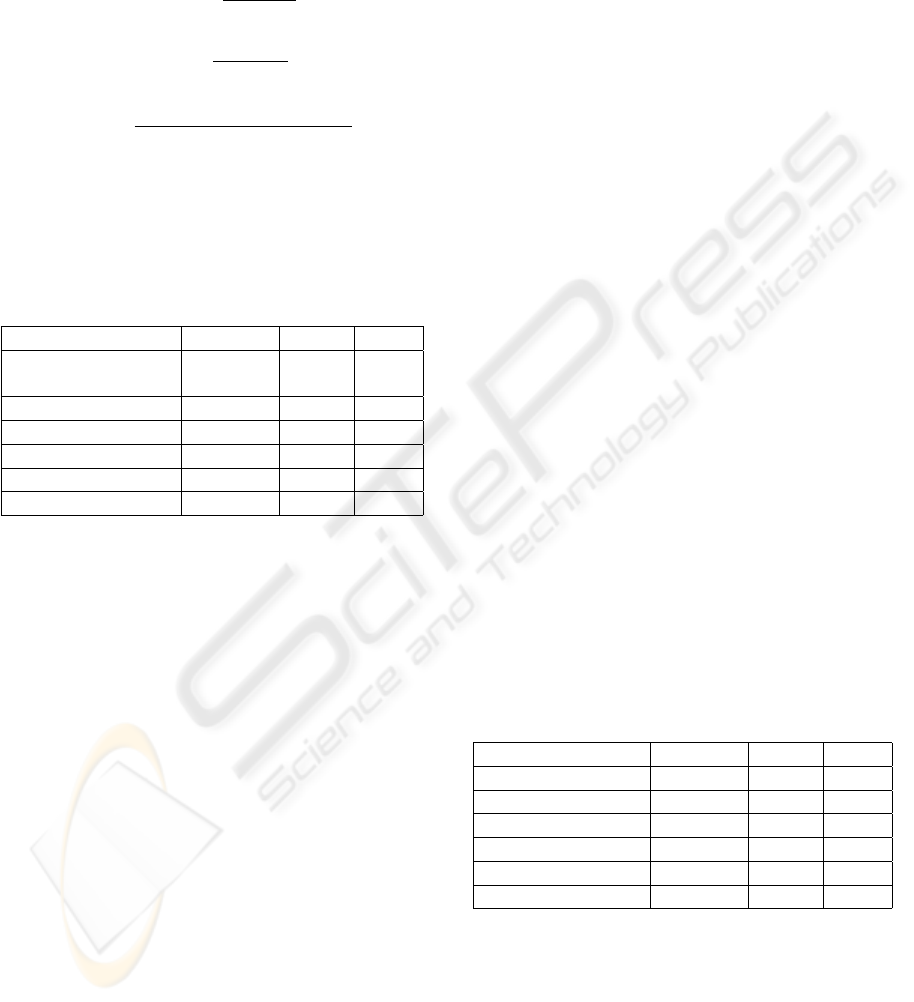
Table 1: Orthographic features.
Feature Example
DigitNumber 12
GreekAlphabet gamma
SingleCapital T
InitialCapital Rho, Cdk
AllCapitals SCK
AllLowers kinin
Hyphen -
Backslash /
LeftBracket [
RightBracket ]
Colon :
SemiColon ;
Percent %
LeftParenthesis (
RightParenthesis )
Comma ,
Period .
Article the, a, an
Conjunction and, or
RomanNumber I, IV, X
LowerMixCapital lacZ
AlphabetMixDigit dig2
Other $, ?
Therefore each token can be represented by the
above 9 features. Furthermore, by sliding windows,
i.e., taking into account a few tokens before and after
the current token, we can retrieve the relevant infor-
mation from the neighboring tokens surrounding the
current token.
In the second layer, we only use one feature, the
predicted class labels from the first layer, and also
make use of sliding windows.
2.5 Post Processing
In order to improve the performance further, we em-
ploy a post-processing module called a boundary
check module, which can recover the following kinds
of boundary errors.
• Boundary errors caused by our BIO represen-
tation: By identifying the label B, the system
can determine the beginning of the named entity.
However, sometimes the system fails to recognize
the label B. So we develop some rules to recover
the label B based on the neighboring tokens. For
example, a sample phrase “maternal alpha feto-
proteins” is recognized into “O maternal”, “I al-
pha” and “I fetoproteins”. According to the labels
of “alpha” and “fetoproteins”, the label of “mater-
nal” is reassigned to “B”.
• Boundary errors due to punctuation: -, +, /, ., etc.
For instance, a phrase “c-myc protein” is recog-
nized into “O c”, “B -”, “I myc” and “I protein”.
In order to recover the integrity, the labels of “c”
and “-” need to be changed into “B” and “I”, re-
spectively.
• Boundary errors due to missing trigger words:
Depending on the annotation guidelines of
BioCreAtIvE challenge, some words need to con-
sidered as part of names, e.g., receptor, enhancer,
promoter, mutant, etc. Hence, for example, if
only “FSH” in “FSH receptor” is recognized as
a named entity, we will add “receptor” into it.
3 RESULTS
The final system is trained on the whole data (15000
sentences). We preform a 5-fold cross validation us-
ing 80% of the data as training data and the remaining
as test data. For the parameter selection, we try a wide
range of SVM cost parameterC from 2
−10
to 2
10
. And
in order to get the most suitable window size in slid-
ing windows, [−le ft, right] window ranges, [−3, 3],
[−2, 2], [−1, 1] and [0, 0] are carried out.
NAMED ENTITY RECOGNITION IN BIOMEDICAL LITERATURE USING TWO-LAYER SUPPORT VECTOR
MACHINES
41
NER systems are often evaluated by means of the
precision, recall and balanced F
β=1
score, which are
defined as follows. “TP”, “FP” and “FN” are the
numbers of true positives, false positives and false
negatives.
Precison =
TP
TP+ FP
Recall =
TP
TP+ FN
F
β=1
=
(β
2
+ 1) · Recall · Precision
β
2
· Recall + Precision
Next, we will present our results and compare
them to other methods that have been applied on the
mostly similar data set, which are shown in Table 2 as
follows.
Table 2: Evaluation results of our system compared with
other systems.
Method Precision Recall F
β=1
Our system
0.835 0.808 0.821
Two-layers SVMs
PosBIOTM-Ner 0.800 0.685 0.738
YamCha 0.825 0.742 0.781
Rule-based tagger 0.843 0.718 0.775
HMM-based tagger 0.803 0.814 0.809
PowerBioNE 0.820 0.832 0.826
Our best result is obtained when C = 1 and
[−le ft, right] window size is [−2, 2]. The results of
PosBIOTM-Ner, Yamcha, Rule-based tagger, HMM-
based tagger and PowerBioNE are taken from (Song
et al., 2004), (Mitsumori et al., 2005), (Tamames,
2005), (Kinoshita et al., 2005) and (Zhou et al., 2005)
respectively.
PosBIOTM-Ner and Yamcha are both based on
SVM algorithm and BIO representation, the same
as our system. PosBIOTM-Ner makes use of edit-
distance as a significant contributing feature for SVM
while Yamcha uses an external lexicon composed of
SWISS-PROT and TrEMBL data. From Table 2, it
can be easily seen that our system based on two-layer
SVMs can outperform these two systems, i.e., F
β=1
score is 8.3% higher than PosBIOTM-Ner and 4%
higher than Yamcha. Also our system can achieve
better performance than Rule-based and HMM-based
taggers.
Notice that our system performs a little worse
than PowerBioNE. We’ve achieved 1.5% higher pre-
cision but 2.4% lower recall, which leads to 0.5%
lower F
β=1
score. This is not surprising for us be-
cause PowerBioNE is a complex system, which is
an ensemble of classifiers in which three classifiers,
one SVM and two discriminative HMMs, are com-
bined using a simple majority voting strategy. In addi-
tion, PowerBioNE incorporates three post-processing
modules to improve the performance further. In the
paper (Zhou et al., 2005), they report that SVM clas-
sifier can get higher precision while HMM classifier
can obtain higher recall. We think that is why our
system has lower recall than PowerBioNE. However,
considering the simplicity of our approach, our sys-
tem is still comparable to PowerBioNE.
In a word, our system can outperform most of
above systems and achieve an approximate state of
the art result, though it is relatively simple and doesn’t
utilize any external lexical resources.
4 DISCUSSION
In the following sections, we start by discussing our
system in detail from different kinds of aspects. Then
we will give a comprehensive error analysis.
4.1 Contributions of Different Features
The effects of each feature in the first layer in our sys-
tem are shown in Table 3. The row of “token(base)”
shows the result when only the token feature is used
in the SVM learning. The other rows show the re-
sults when the token feature plus one other feature are
used in the learning, i.e.,“+orthographic” means the
token plus the orthographic feature; “+POS” means
the token feature plus the MedPost POS feature; “+bi-
prefix/suffix” means the token feature plus the bi-
prefix and bi-suffix features, etc.
Table 3: Contributions of each feature after added into the
base feature.
Precision Recall F
β=1
token (base) 0.714 0.491 0.581
+orthographic 0.729 0.709 0.719
+MedPost POS 0.717 0.541 0.616
+bi-prefix/suffix 0.760 0.615 0.680
+tri-prefix/suffix 0.750 0.571 0.648
+quad-prefix/suffix 0.728 0.530 0.613
From Table 3, it shows that the orthographic fea-
ture and the bi-prefix/suffix feature are critical for
our system, which improve F
β=1
score by 13.8% and
9.9% compared with the base. Moreover, most of the
features in Table 3 can’t affect the precision so much
while they mostly have great effects on the recall.
Then, as mentioned above, we use MedPost for
POS tagging, which was trained on the MEDLINE
ICEIS 2007 - International Conference on Enterprise Information Systems
42
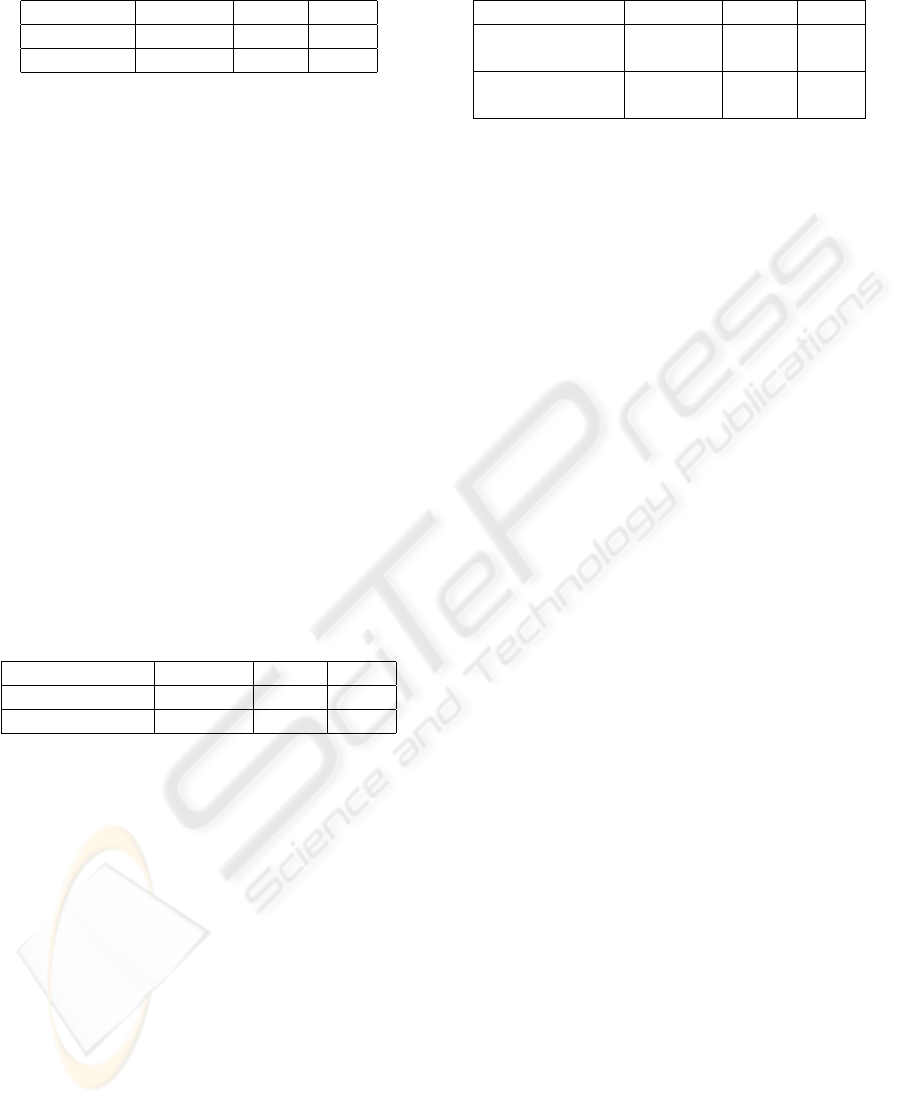
Table 4: Effects of different POS taggers.
POS tagger Precision Recall F
β=1
MedPost 0.770 0.783 0.777
TreeTagger 0.764 0.774 0.769
corpus. To demonstrate the effects of using differ-
ent POS taggers in our system, we replace MedPost
with TreeTagger (Schmid, 1994), which was trained
on newswire articles, to retrain our system. The re-
sults are shown in Table 4. It can be seen that MedPost
performs slightly better than TreeTagger but the dif-
ference is too small to be significant. Together with
the results from Table 3, we can conclude that the
POS feature can’t affect our system very much.
4.2 Contributions of the Second Layer
Introducing a second layer isn’t unusual. A similar
approach has been applied successfully in protein sec-
ondary structure prediction (Rost, 2003).
Our system performs prediction in two layers,
where the first layer is a text to entity step and the
second layer is a entity to entity step, i.e., the named
entities that are predicted in the first layer are fed to a
new classifier in the second layer. Table 5 shows the
detailed results of various layers in our system.
Table 5: Contributions of various layers in our system.
Precision Recall F
β=1
the first layer 0.770 0.783 0.777
the second layer 0.811 0.792 0.801
From Table 5, it is clear that the precision, recall
and F
β=1
score are increased by 4.1%, 0.9% and 2.4%
respectively, which means that the second layer can
improve the performance of our system greatly. We
think the reason is that based on the prior prediction
from the first layer and making use of sliding win-
dows, our system can output more refined named en-
tities according to the prior prediction together with
its neighbors.
4.3 Effect of Post Processing
We carry out a boundary check post-processing mod-
ule to make sure the integrity of the named entities.
Table 6 shows the performance of the boundary check
module.
In NER systems, boundary errors of named enti-
ties are very critical because they can increase both
false positive errors and false negative errors. We
can elaborate it by the following example. Suppose
Table 6: Performance of the boundary check module.
Precision Recall F
β=1
before
0.811 0.792 0.801
boundary check
after
0.835 0.808 0.821
boundary check
that a sample phrase “NF-kappa Bp50” is recognized
wrongly to “O NF”, “B -”, “I kappa” and “I Bp50”,
which leads to both a false positive error (“-kappa
Bp50” isn’t a gene/protein name) and a false neg-
ative error (“NF-kappa Bp50” isn’t recognized cor-
rectly). Therefore, it is essential to introduce a bound-
ary check module in our system.
From Table 6, it is clear that the precision, re-
call and F
β=1
score are increased by 2.4%, 1.6% and
2.0%, respectively. This means that the boundary
check module can effectively correct some boundary
errors and consequently improve the performance of
our system further.
4.4 Error Analysis
Finally, we conduct an error analysis to find out where
our system can be improved in the future. We catego-
rize all the errors into 5 classes.
• boundary errors only occur on the left side, which
are those errors with the correct right boundary
but the incorrect left boundary.
• boundary errors only occur on the right side,
which are those errors with the correct left bound-
ary but the incorrect right boundary.
• boundary errors occur on both sides, which are
those errors with both the wrong right boundary
and the wrong left boundary.
• non-boundary false positive errors are those errors
which are wrongly recognized into named entities
and don’t contain any boundary errors.
• non-boundary false negative errors are those er-
rors which are missed being identified to named
entities and don’t contain any boundary errors.
Figures 2 and 3 show the distribution of the above
5 kinds of errors before and after a boundary check
module, respectively.
Boundary errors may be the easiest ones to solve
because at least our system has correctly recognized
part of the named entities, which can give useful clues
to correct the errors. Through comparing Figures 2
and 3, the boundary check module indeed can reduce
some boundary errors, about 2% less on average. But
NAMED ENTITY RECOGNITION IN BIOMEDICAL LITERATURE USING TWO-LAYER SUPPORT VECTOR
MACHINES
43

Figure 2: Proportions of different classes of errors before a
boundary check module.
Figure 3: Proportions of different classes of errors after a
boundary check module.
we need optimize this module further since there are
still 27% boundary errors left in our system.
The remaining 73% errors are composed of false
positive errors (33%), which often occur when a noun
phrase is a generic term (“mRNA”, “gene”) or mixed
with digits and capitals (“W85”), and false negative
errors (40%), which often occur when our system has
little related information and context clues.
5 CONCLUSION
In this paper, we first propose a named entity recog-
nition system for biomedical literature using two-
layer support vector machines and then present a post-
processing module called a boundary check module
to improve the performance further. The results show
that with carefully designed features and introducing
a second layer, our system can recognize named enti-
ties with fairly high accuracy, even without using ex-
ternal lexicons.
In the future, we will consider more modules to re-
solve the abbreviations, the aliases, etc. Furthermore,
we will explore appropriate approaches to make use
of external knowledge resources, e.g., lexicons, full
MEDLINE abstracts and web searches.
ACKNOWLEDGEMENTS
The first author Feng Liu is funded by a doctoral grant
of the Vrije Universiteit Brussel (VUB) and the sec-
ond author Yifei Chen is funded by a doctoral grant of
the Fonds voor Wetenschappelijk Onderzoek (FWO).
REFERENCES
BioCreAtIvE (2006). The second biocreative - critical as-
sessment for information extraction in biology chal-
lenge. http://biocreative.sourceforge.net.
Chang, C.-C. and Lin, C.-J. (2001). LIBSVM: a library
for support vector machines. Software available at
http://www.csie.ntu.edu.tw/ cjlin/libsvm.
Kinoshita, S., Cohen, K. B., Ogren, P. V., and Hunter, L.
(2005). Biocreative task 1a: entity identification with
a stochastic tagger. BMC Bioinformatics, 6(Suppl
1):S4.
MEDLINE (1966). http://www.nlm.ih.gov.
Mitsumori, T., Fation, S., Murata, M., Doi, K., and Doi,
H. (2005). Gene/protein name recognition based on
support vector machine using dictionary as features.
BMC Bioinformatics, 6(Suppl 1):S8.
Morgan, A. A., Hirschman, L., Colosimo, M., Yeh, A. S.,
and Colombe, J. B. (2004). Gene name identification
and normalization using a model organism database.
Journal of Biomedical Informatics, 37(6):396–410.
Rost, B. (2003). Rising accuracy of protein secondary struc-
ture prediction. Protein structure determination, anal-
ysis, and modeling for drug discovery, pages 207–249.
Schmid, H. (1994). Probabilistic part-of-speech tagging us-
ing decision trees. In International Conference on
New Methods in Language Processing, Manchester,
UK.
Smith, L., Rindflesch, T., and Wilburn, W. J. (2004). Med-
post: a part-of-speech tagger for biomedical text.
BIOINFORMATICS, 20(14):2320–2321.
Song, Y., Yi, E., Kim, E., and Lee, G. G. (2004). Posbiotm-
ner: A machine learning approach for bio-named en-
tity recognition. In Proceedings of BioCreAtIvE Work-
shop, Granada, Spain.
Takeuchi, K. and Collier, N. (2003). Bio-medical entity ex-
traction using support vector machine. In Proceedings
of the ACL-03 Workshop on Natual Language Pro-
cessing in Biomedicine, pages 57–64.
ICEIS 2007 - International Conference on Enterprise Information Systems
44

Tamames, J. (2005). Text detective: a rule-based system for
gene annotation in biomedical texts. BMC Bioinfor-
matics, 6(Suppl 1):S10.
Tanabe, L. and Wilbur, W. J. (2002). Tagging gene and
protein names in biomedical text. Bioinformatics,
18(8):1124–1132.
Tsuruoka, Y. and Tsujii, J. (2003). Boosting precision and
recall of dictionary-based protein name recognition.
In Proceedings of the ACL 2003 Workshop on NLP in
Biomedicine, pages 41–48.
Vapnik, V. (1995). The nature of statistical learning theory.
Springer-Verlog, New York.
Zhou, G., Shen, D., Zhang, J., Su, J., and Tan, S. (2005).
Recognition of protein/gene names from text using an
ensemble of classifiers. BMC Bioinformatics, 6(Suppl
1):S7.
NAMED ENTITY RECOGNITION IN BIOMEDICAL LITERATURE USING TWO-LAYER SUPPORT VECTOR
MACHINES
45
