
BREAST CANCER DETECTION USING GENETIC
PROGRAMMING
Hong Guo, Qing Zhang and Asoke K. Nandi
Department of Electrical Engineering and Electronics
The University of Liverpool, Brownlow Hill, Liverpool, L69 3GJ, U.K.
Keywords:
Genetic Programming, Feature Extraction, Classification, Breast Cancer Diagnosis.
Abstract:
Breast cancer diagnosis have been investigated by different machine learning methods. This paper proposes a
new method for breast cancer diagnosis using a single feature generated by Genetic Programming (GP). GP as
an evolutionary mechanism that provides a training structure to generate features. The presented approach is
experimentally compared with some kernel feature extraction methods: Kernel Principal Component Analysis
(KPCA) and Kernel Generalised Discriminant Analysis (KGDA). Results demonstrate the capability of the
proposed method to transform information from high dimensional feature space into one dimensional space
for breast cancer diagnosis.
1 INTRODUCTION
Breast Cancer is the second most common cancer in
the UK after non-melanoma skin cancer (Can). The
early detection of breast cancer is becoming very im-
portant to ameliorate breast cancer survival rate. In
recent years, various machine learning methods have
been proposed for breast cancer diagnosis and prog-
nosis. Yao and Liu described two neural network
based approaches to breast cancer diagnosis; a feed-
forward neural networks was evolved using evolu-
tionary programming algorithm in the first approach,
while the second approach was based on neural net-
work ensembles (Yao and Liu, 1999). The perfor-
mance of four fuzzy rule generation methods on Wis-
consin breast cancer data was studied in (Jain and
Abraham, 2004). In (Kermani et al., 1995), a hy-
brid genetic algorithm and neural network (GANN)
was shown to extract the important features and train
a NN in breast cancer classification. Guo and Nandi
developed a modified Fisher criterion to help genetic
programming optimism features for breast cancer di-
agnosis (Guo and Nandi, 2006). Nandi et al. used
GP successfully for classification of breast masses in
mammogram (Nandi et al., 2006).
In recent years, the application of genetic pro-
gramming to pattern recognition problem has become
increasingly common. Genetic Programming was
first introduced by Koza (Koza, 1992), and has been
proposed as a machine learning method in different
fields. In (Benyahia and Potvin, 1998), GP technique
was used to develop a decision support system for ve-
hicle dispatching considering a population of utility
functions that evaluate candidate vehicles for servic-
ing requests. GP was tested in six medical diagno-
sis problems (Brameier and Banzhaf, 2001) and the
results were compared with those obtained by neu-
ral networks. In (Kishore et al., 2000) the feasibility
of applying GP to multi-category pattern classifica-
tion problem was studied. Zhang et al. (Zhang et al.,
2003) applied genetic programming for fault detec-
tion in machine condition monitoring field. However,
in all the above applications (Benyahia and Potvin,
1998; Brameier and Banzhaf, 2001; Kishore et al.,
2000; Zhang et al., 2003), GP was employed solely
as a classifier based on manually developed features.
In (Sherrah et al., 1997), GP-based feature extraction
was used to improve the classification results and re-
duce the dimensionality of the data in the medical
domain. GP exhibits pseudo-intelligent behaviour by
deciding whether to perform feature extraction or fea-
ture selection during the evolutionary process. Unfor-
tunately, the system is unable to sample adequately
the search space for high-dimensional problems and
334
Guo H., Zhang Q. and K. Nandi A. (2008).
BREAST CANCER DETECTION USING GENETIC PROGRAMMING.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 334-341
DOI: 10.5220/0001059203340341
Copyright
c
SciTePress

the main disadvantage lies in its computational com-
plexity. Kotani et al. (Kotani et al., 1997) performed
feature extraction using GP with a KNN classifier on
one artificial task and one acoustic diagnosis exper-
iment with the conclusion that the genetic program-
ming is an effective tool for the feature extraction
task.
In this paper, GP is employed to generate a single
nonlinear feature to improve the classification accu-
racy for breast cancer diagnosis. As a machine learn-
ing method, GP exhibits intelligent behaviour to per-
form feature generation. During the evolutionary pro-
cess, a new fitness function is developed to evaluate
the effectiveness of each feature in helping GP select
the best features by which the patterns from benign
are well separated from patterns from malignant.
This paper is organized as follows: The data
preparation of breast cancer is addressed in Section
2. Section 3 presents the proposed feature generator
using genetic programming. Two kernel feature ex-
traction methods kernel principal component analysis
(KPCA) and kernel generalized discriminant analy-
sis (KGDA) are briefly presented in section 4. Three
classifiers Multi-Layer Prceptron (MLP), k-Nearest
Neighbor (KNN) and Minimum Distance Classifier
(MDC) are presented in section 5. In section 6, a num-
ber of experiments for breast cancer detection prob-
lems are reported using kernel Principal Component
Analysis, kernel Generalized Discriminant Analysis
extracted features and GP generated feature. Finally,
based on the experimental results, conclusions on this
proposed method are presented in section 7.
2 THE PROBLEM
It is of prime importance to be able to detect the breast
cancer in early stages. In this paper, the Wisconsin
diagnostic breast cancer (WDBC) dataset from the
UCI Machine Learning repository (D.J. Newman and
Merz, 1998) is used to examine the capability of GP
for the breast cancer detection problem.
2.1 Image Preparation
The Wisconsin diagnostic breast cancer (WDBC)
dataset was created by Wolberg et al., University of
Wisconsin (Street et al., 1993). The diagnosis proce-
dure begins by obtaining a small drop of fluid from
a breast tumour using a fine needle. The image for
digital analysis is generated by JVC TK-1070 colour
video camera mounted atop an Olympus microscope
and the image is projected into the camera with a
63× objective and a 2.5× ocular. The image is cap-
tured by a ComputerEyes/RT colour frame grabber
board (Digital Vision, Inc., Dedham MA 02026) as
a 512× 480, 8-bit-per-pixel Targa file.
2.2 Data Preparation
An active model located in the actual boundary of cell
nucleus is defined as a snake. The ten different fea-
tures from the snake-generated cell nuclei boundaries
are extracted by following techniques:
• Radius: The radius of an individual nucleus is
measured by averagingthe length of the radial line
segments defined by the centroid of the snake and
the individual snake points.
• Perimeter: The nuclear perimeter is defined by
calculating the total distance between the snake
points.
• Area: The nuclear area is defined by counting the
number of pixels on the interior of the snake and
adding one-half of the pixels in the perimeter.
• Compactness: The perimeter
2
/area is used as
the compactness of the cell nuclei.
• Smoothness: The smoothness of a nuclear con-
tour is quantified by measuring of difference be-
tween the length of a radial line and the mean
length of the lines surrounding it.
• Concavity: Concavity is defined as the severity of
indentations in a cell nucleus. For a line connect-
ing any two non-adjacent snake points, if the ac-
tual boundary drop inside the line, an indentation
occurs and the distance to the line is a measure of
the severity.
• Concave Points: This feature is similar to Con-
cavity but measures only the number, rather than
the magnitude, of contour concavities.
• Symmetry: The length difference between lines
perpendicular to the major axis to the cell bound-
ary in both directions is defined as symmetry.
• Fractal Dimension: The fractural dimension is an
indication of the regularity of the nucleus. Higher
values of the downward slops of the coastlines
correspond to less regular contour and vice-versa.
• Texture: The texture of the cell nucleus is defined
by finding the variance of the gray scale intensities
in the component pixels.
BREAST CANCER DETECTION USING GENETIC PROGRAMMING
335

TRoot
tanh
feature2
feature1
Figure 1: Tree Representation.
The mean value, largest value and standard error of
each feature are computed for each image. A set of
569 images has been processed, yielding a database of
30-dimensional points (Street et al., 1993). In this pa-
per, we randomly selected, without replacement, 100
samples for benign case, and 100 samples for malig-
nant case respectively. Two 30× 200 matrices are ob-
tained for training and test datasets. One of them as
the training dataset forms the terminator set to the GP.
Another matrix is used as the test dataset. For each
given pattern vector of training and test datasets, a
corresponding vector is created in a matrix contain-
ing the target information.
3 GENETIC
PROGRAMMING-BASED
FEATURE GENERATOR
In this paper, we introduce a new method for a feature
generator based on GP, for breast cancer detection
problem. Genetic Programming, as a form of evo-
lutionary algorithm and an extension of genetic algo-
rithms, extracts the information from the real-valued
parameter vector to create features based on the evo-
lutionary algorithm. The surviving feature from the
feature generator will be used to provide the solution
to pattern recognition problems.
3.1 The Representation of Each
Individual
Since expressions can be represented as trees or-
dered by operator precedence, GP systems in this pa-
per evolve programs using tree representation. Each
member can be written as a polynomial expression
consisting of several non-linear functions up to a max-
imum specified depth. Using this function, each in-
dividual in the population is a mathematical formula
that transforms the time series signals into a feature
data. Formula TRoot = tanh( feature1) + feature2
can be represented by the Fig. 1.
3.2 Process of Genetic Programming
The GP-based feature extractor is used to extract use-
ful information from the thirty features of breast can-
cer dataset in order to provide discriminating input
features for the classifiers. The purpose of GP is to try
to maximise the extra information content in the sam-
ple of the original feature set, and it implicitly max-
imises the separation between benign condition and
malignant condition within the data. The evolution-
ary process of GP-based feature generation system is
described by following steps. First, an initial popula-
tion with a chosen number of individuals is generated
on a random basis, meaning that there is no human in-
fluence or bias in the generation of original features.
Original feature set are fed as the inputs to the ini-
tial population. Each individual represents a transfor-
mation network, which tries to transform dataset into
information for classification.
In terms of the usefulness of each individual for
classification, a fitness value is assigned to each in-
dividual by fitness function. The members with the
best fitness values survive from the current genera-
tion and will be chosen as the origins of the next gen-
eration. In our design, only the elite will survive the
natural selection. This mechanism allows the feature
to evolve in a direction towards the best classification
performance, thus achieving the automatic generation
of features. At the beginning of the next generation,
three operations - reproduction, crossover and muta-
tion - are conducted to produce new members based
on the surviving member. If the termination criterion
is met, the best solution is preserved.
3.3 Fitness Function
The fitness function is one of the most important com-
ponents. It determines the performance of the GP sys-
tem. A good fitness function provides an improved
solution by rating the performance of each member
and giving the stronger one a better chance of surviv-
ing. It is well known that the computational demands
are relatively high in training a classifier for each in-
dividual when the classification results are used as
the fitness value for breast cancan diagnosis problem.
Hence in this study it is decided that classification re-
sults are not used as a measure of fitness. This deci-
sion reduces the computational complexity of the pro-
posed method significantly
Within the one-dimensional effective feature
space, the achievable classification success is de-
pendent upon the overlapping areas between classes.
Usually, a threshold is set within the area to sepa-
rate data belonging to different classes. However, it
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
336

Figure 2: Crossover operation
is inevitable that some data points are misclassified.
Apparently the smaller the overlapping area, by the
smaller number of data points within the overlapping
area, the higher is the classification success. This rule
is explored in our fitness function to reveal quickly
and effectively the discriminating ability of the candi-
date features. Specifically, the higher boundary of the
lower class and the lower boundary of the higher class
are calculated. The number of data points present
within these two boundaries are found out and then
normalised by the total number of data points. A
small percentage of the overlapping points is an in-
dication how well the two classes can be separated.
3.4 Primitive Operations
Genetic programming evolves tree individuals repre-
senting possible solutions to the problem at hand. A
population of such individuals is randomly created
and then evolved by probability of genetic operations:
• Crossover: GP carries out a crossover operation
to create new individuals with a probability P
c
,
which controls the occurrence of the crossover
throughout generations. Two new individuals
are generated by selecting compatible nodes ran-
domly from each parent and swapping them, as
illustrated in Fig. 2.
• Mutation: The mutation operation is performed
by the creation of a subtree at a randomly selected
node with the probability P
m
. First, for a given
parent, there is an index assigned to each node
for identification. A random index number is gen-
erated to indicate the place where mutation will
happen. The node is located, then the tree down-
stream from this node is deleted and a new subtree
is generated from this node (see Fig. 3), exactly
in the same way as growing initial population.
• Reproduction: The reproduction operation is per-
formed by copying individuals to the next pop-
ulation without any change in terms of a certain
probability P
r
.
Figure 3: Mutation operation.
Table 1: The Operator sets for the GP.
Symbol No. of Inputs Description
+, - 2 Addition, Subtraction
*, / 2 Multiplication, Division
square, sqrt 1 Square, Square Root
sin, cos 1 Trigonometric functions
asin, acos 1 Trigonometric functions
tan, tanh 1 Trigonometric functions
reciprocal 1 Reciprocal
log 1 Natural Logarithm
abs, negator 1 Absolute, Change Sign
All these three operations happen within one gen-
eration based on three probabilities, such that:
P
c
+ P
m
+ P
r
= 1 (1)
3.5 Primitive Terminators
Terminators act as the interface between GP and the
experimental dataset. They are required to collect re-
lated information as much as possible from the orig-
inal feature set and to provide inputs to the feature
generator. In our GP-based feature extractor, the ter-
minator set is constructed by thirty original feature set
(see Section 2) and some numerical values, which are
randomly generated at the construction cycle of new
individuals. These numerical values could be either
integer or floating point numbers, both ranging from
1 to 100.
3.6 Primitive Operators
One of the main building blocks of the GP is the oper-
ator pool. The functions stored in the pool are math-
ematical operators that perform an operation on one
or more inputs to give an output result. Table 1 lists
the mathematical functions used as operators in this
paper.
BREAST CANCER DETECTION USING GENETIC PROGRAMMING
337

4 KERNEL FEATURE
EXTRACTION METHODS
In recent years, kernel-based methods are becoming
popular for their ability to solving nonlinear prob-
lems. It is first applied to overcome the computational
and statistical difficultly of SVM classifier for seek-
ing an optimal separating hyperplane in the feature
space(E.Osuna et al., 1997). It is demonstrated to be
able to represent complicated nonlinear relationship
of the input data efficiently.
The Kernel Principal Component Analysis
(KPCA) and Kernel Generalised Discriminant
Analysis (KGDA) are two independent nonlinear
feature extraction/selection methods, both of which
perform the mapping in the feature space F with
kernel functions and use a linear analysis algorithm
to discover patterns in the kernel-defined space.
The mapping function Φ is defined implicitly by
specifying the form of the dot product in the feature
space (Scholkopf et al., 1998).
4.1 Kernel Principal Component
Analysis
Kernel PCA is the non-linear extension of the PCA in
a kernel-defined feature space making use of the dual
representation (Shawe-Taylor and Cristianini, 2004).
Given a set of observations {~x
i
∈ R
n
: i = 1 to N},
we first map the data into a feature space F and com-
pute the covariance matrix(Muller et al., 2001):
C =
1
N
N
∑
j=1
Φ(x
j
)Φ(x
j
)
T
(2)
The N × N Kernel Matrix is defined as,
K
ij
:= Φ(~x
i
) • Φ(~x
j
) = K(~x
i
,~x
j
); i, j = 1,...,N
(3)
The data need to be centred in the mapped feature
space F
˜
K
ij
≡
˜
Φ(~x
i
) •
˜
Φ(~x
j
) = K
ij
−
1
N
N
∑
p=1
K
ip
−
1
N
N
∑
q=1
K
qj
+
1
N
2
N
∑
p,q=1
K
pq
(4)
Now the eigenvalue problem for the expansion coef-
ficients α
i
is solely dependent on the kernel function,
λα =
˜
Kα (5)
Projects the mapped pattern Φ(x) onto V
k
to extract
features of new dataset x with kernel PCA.
(V
k
· Φ(x)) =
N
∑
i=1
α
k
i
(Φ(x
i
) · Φ(x)) =
N
∑
i=1
α
k
i
K(x
i
,x)
(6)
4.2 Kernel Generalized Discriminant
Analysis (KGDA)
KGDA is derived from a linear version of the dis-
criminant analysis, namely, Fisher linear discriminant
analysis FLDA. FLDA is designed optimally with its
ability to maximise the ratio of within-class scatter
and between-class scatter of projected features. For
c (c > 2) classes, the ith observation vector from the
class l is defined by x
li
, where 1 ≤ l ≤ c, 1 ≤ i ≤ N
l
,
and N
l
is the number of observationsfrom class l. The
within-class covariance matrix is given by
S
ω
=
c
∑
l=1
S
l
, (7)
where
S
l
=
N
l
∑
i=1
(x
li
− µ
l
)(x
li
− µ
l
)
T
(8)
The between-class covariance matrix is defined by
S
b
=
c
∑
l=1
N
l
(µ
l
− µ)(µ
l
− µ)
T
(9)
where µ
l
is the mean of class l and µ is the global
mean.
The idea of KGDA is to solve the problem of
FLDA in a kernel feature space, thereby yielding a
nonlinear discriminant in the input space. In term
of the dot product, the optimisation problem for the
KGDA in the feature space can be written as
J(α) =
α
T
S
Φ
b
α
α
T
S
Φ
ω
α
(10)
where
S
Φ
b
=
c
∑
l=1
[k
l
k
T
l
− kk
T
] (11)
S
Φ
ω
= K
2
−
c
∑
l=1
N
l
k
l
k
T
l
(12)
k
l
=
1
N
l
N
l
∑
l=1
K
ij
i, j = 1,...,N
l
(13)
k =
1
N
N
∑
i=1
K
ij
i, j = 1,...,N (14)
where k
l
is the mean vector of kernel matrix of class
l, k indicates the global mean vector of kernel matrix
of K
ij
.
The projection of the test dataset x into the dis-
criminant is given by
W · Φ(x) =
N
∑
i=1
α
i
k(x
i
,x) (15)
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
338

5 CLASSIFIERS
Three classifiers - Artificial Neural Networks
(ANNs), K-Nearest Neighbour (KNN) and Minimum
Distance Classifier (MDC) - are employed in this
paper to evaluate the discriminating ability of features
generated by GP and other kernel feature extraction
methods discussed previously.
The Multi-Layer Peceptron (MLP) is chosen here
as the structure of the network for its overall perfor-
mance over other configurations. The MLP used here
consists of one hidden layer varying between 1 and 14
neurons and one output layer, with the hidden layer
having a logistic activation function and the output
layer using a linear activation function. For training
procedure, the back propagation algorithm with adap-
tive learning and momentum is used. The network is
trained for 10000 epochs using each feature set.
KNN is a supervised learning algorithm to classify
a test object based on majority of K-nearest neighbor
category. Given that the version of K = 1 is often
rather successful (Ripley, 2004). 1-NN is used as the
classifier to examine the performance of features in
this paper.
MDC is the simplest classification criterion. Basi-
cally, the method finds centres of classes and mea-
sures distances between these centres and the test
data. The distance is defined as a measure of similar-
ity so that the minimum distance indicates the max-
imum similarity. In this paper, Euclidean distance is
used to investigate the capability of any feature ex-
tracted by this approach.
6 EXPERIMENTAL RESULTS
6.1 Feature Generation Result
Fig. 4 is obtained for detection of breast cancer by
running GP-based feature extractor with population
size 100, maximum tree depth 10 and terminating af-
ter the number of generations reaches 5000. Fig. 4
shows the output of a single feature, generated from
the original feature set with 30 dimensions, for the
training dataset and test dataset respectively. There
are 200 examples in total from two conditions, with
100 examples in the benign case and 100 examples
in the malignant case. It is clear from Figure 4 that
the two conditions are perfectly separated from each
other at training dataset, and three examples misclas-
sified in test dataset.
0 50 100 150 200
−1
−0.5
0
0.5
1
Training data
0 50 100 150 200
−1
−0.5
0
0.5
1
Test data
Figure 4: Output of a single feature, generated by GP from
the original feature set with 30 dimensional breast cancer
data, for the 200 examples in each of the training dataset
and test dataset respectively.
6.2 Classification Results
A number of experiments were carried out to evalu-
ate the discriminating ability of features generated by
GP and other classical feature extraction methods in
term of classification performance using MLP, 1-NN
and the simplest classifier MDC respectively. Twenty
runs of GP has been conducted for generating fea-
tures. Also, fifty MLP have conducted using original
features and feature extracted by KPCA, KGDA and
GP respectively.
Table 2 presents the comparison results of classifi-
cation success rate using feature set extracted by dif-
ferent method as the inputs to MLP, 1-NN and MDC.
It can be seen that the best classification accuracy is
achieved by MLP when thirty original features are
used as input. One KPCA feature achieved the best
94.5% when MDC is use as the classifier. one KGDA
feature with MLP and MDC achieved the same clas-
sification results 93.5%. When a GP extracted fea-
ture is employed, the improvementis significant com-
pared with other classical feature extraction methods.
Together with MLP, KNN and MDC, it performs the
best with success rate at 98.5% among all of pattern
recognition systems. From the best classification ac-
curacy it can be seen that GP generated features are
more robust compared with other methods.
BREAST CANCER DETECTION USING GENETIC PROGRAMMING
339
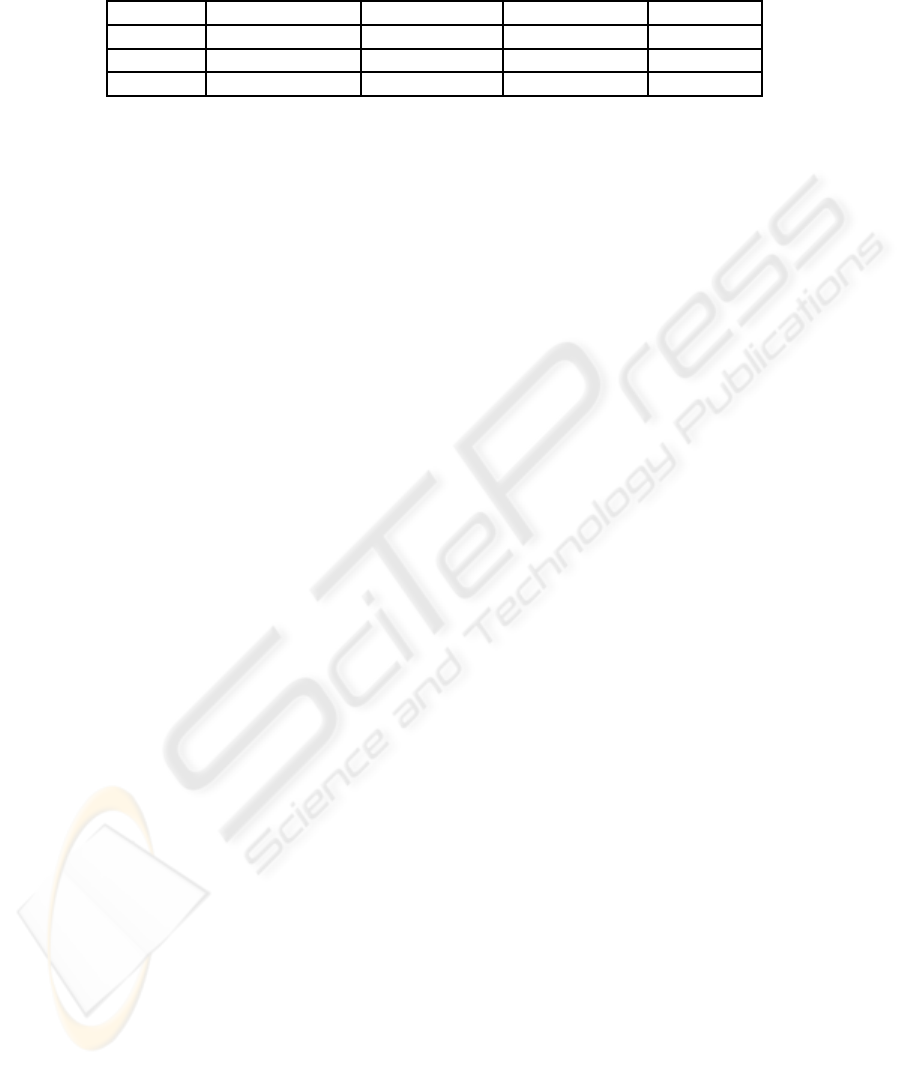
Table 2: The best classification accuracy (%) using original features, one KPCA-extracted features, one KGDA-extracted
features and one GP-generated features respectively, with a MLP, a KNN and a MDC classifier respectively on breast cancer
dataset.
Classifier Original Feature KPCA Feature KGDA Feature GP Feature
MLP 97% 90% 93.5% 98.5%
KNN 87.5% 85.5% 93% 98.5%
MDC 84% 94.5% 93.5% 98.5%
7 CONCLUSIONS
It is now clear from Figure 4 that values of the single
feature obtained from our proposed method cluster
naturally into largely non-overlapping groups. Thus
no computationally complex classifier may be needed
for successful classification, instead some simple
thresholds are enough. Summarizing all the results
obtained from different approaches for breast cancer
diagnosis problem, it can be said that performances
from a single GP-generated feature are the most accu-
rate and reliable in all experiments. From the results
of different pattern recognition problems, GP is not
only capable of reducing the dimensionality, but also
achieving a significant improvement in the classifica-
tion accuracy. Using the single feature generated by
GP makes a significant contribution to the improve-
ment in classification accuracy and robustness, com-
pared with other sets of features extracted by KPCA
and KGDA.
Generally in pattern recognition problems, there is
a reliance on the classifier to find the discriminating
information from a large feature set in case of stand-
alone MLP. In this paper, GP as a machine learning
method is proposed for nonlinear feature extraction
for breast cancer diagnosis. This approach is able
to learn directly from the data just like conventional
methods (such as FLDA and PCA), but in an evolu-
tionary process. Under this framework, an effective
feature can be formed for pattern recognition prob-
lems without the knowledge of probabilistic distribu-
tion of data.
From the experimental results it can be seen that
with the combination of a simple form of classifier
MDC, GP outperforms the other two feature extrac-
tors which are using more sophisticate classifier MLP,
indicating an overwhelming advantage of GP in fea-
ture extraction for breast cancer diagnosis.
ACKNOWLEDGEMENTS
H. Guo would like to acknowledge the financial sup-
port of the Overseas Research Studentship Commit-
tee, UK, the University of Liverpool and the Univer-
sity of Liverpool Graduates Association (HK)
REFERENCES
Cancer research UK.
Benyahia, I. and Potvin, J. (1998). Decision support for
vehicle dispatching using genetic programming. IEEE
Trans. Syst., Man, Cybern. Part.A, 28(3):306–314.
Brameier, M. and Banzhaf, W. (2001). A comparison of
linear genetic programming and neural networks in
medical data mining. IEEE Trans. on Evolutionary
Computation, 5(1):17–26.
D.J. Newman, S. Hettich, C. B. and Merz, C. (1998). UCI
repository of machine learning databases.
E.Osuna, Freund, R., and Girosi, F. (1997). Support Vec-
tor Machines: Training and Applications. MIT, Tech.
Rep.
Guo, H. and Nandi, A. K. (2006). Breast cancer diagnosis
using genetic programming generated feature. Pattern
Recognition, 39(5):980–987.
Jain, R. and Abraham, A. (2004). A comparative study
of fuzzy classification methods on breast cancer data.
Australas. Physical Engineering Sciences Medicine,
27(4):213–218.
Kermani, B. G., White, M. W., and Nagle, H. T. (1995).
feature extraction by genetic algorithms for nerual net-
works in breast cancer classification. volume 1, pages
831–832. New York, USA.
Kishore, J. K., Patnaik, L. M., Mani, V., and Arawal, V. K.
(2000). Application of genetic programming for mul-
ticategory pattern classification. IEEE Trans. on Evo-
lutionary Computation, 4(3):242–258.
Kotani, M., Ozawa, S., Nasak, M., and K.Akazawa (1997).
Emergence of feature extraction function using ge-
netic programming. In Knowledge-Based Intelligent
Information Engineering Systems, Third International
Conference, pages 149–152.
Koza, J. R. (1992). Genetic Programming: On the Pro-
gramming of Computers by Means of Natural Selec-
tion. MIT Press, Cambridge.
Muller, K. R., Mika, S., Ratsch, G., Tsuda, K., and
Scholkopf, B. (2001). An introduction to kernel-based
learning algorithms. IEEE Trans. on Neural Networks,
12(2):181–201.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
340

Nandi, R. J., Nandi, A. K., Rangayyan, R. M., and Scutt,
D. (2006). Classification of breast masses in mammo-
grams using genetic programming and feature selec-
tion. Medical and Biological Emgineering and Com-
puting, 44(8):693–694.
Ripley, B. D. (2004). Pattern Recognition and Neural Net-
works. Cambridge Universith Press, Boston.
Scholkopf, B., Smola, A., and Muller, K. R. (1998). Non-
linear component analysis as a kernel eigenvalue prob-
lem. Neural Computation, 10(5):1299–1319.
Shawe-Taylor, J. and Cristianini, N. (2004). Kernel Meth-
ods for Pattern Analysis. Cambridge University Press.
Sherrah, J. R., Bogner, R. E., and Bouzerdoum, A. (1997).
The evolutionary pre-processor: Automatic feature
extraction for supervised classification using genetic
programming. In Proc. 2nd Int. Conf. Genetic Pro-
gramming (GP-97), pages 304–312.
Street, W., Wolberg, W., and Mangasarian, O. (1993). Nu-
clear feature extraction for breast tumor diagnosis. In
International Symposium on Electronic Imaging: Sci-
ence and Technology, number 1905, pages 861–870.
San Jose, CA.
Yao, X. and Liu, Y. (1999). Neural networks for breast can-
cer diagnosis. volume 3, pages 6–9.
Zhang, L., Jack, L. B., and Nandi, A. K. (2003). Fault detec-
tion using genetic progamming. Mechanical Systems
Signal Processing.
BREAST CANCER DETECTION USING GENETIC PROGRAMMING
341
