
FEASABILITY OF YEAST AND BACTERIA IDENTIFICATION
USING UV-VIS-SWNIR DIFUSIVE
REFLECTANCE SPECTROSCOPY
J. S. Silva, R. C. Martins, A. A. Vicente and J. A. Teixeira
IBB - Institute for Biotechnology and BioEngineering, Universidade do Minho
Campus de Gualtar, 4710-057 Braga, Portugal
Keywords:
Yeast, bacteria, UV-VIS-SWNIR reflectance spectroscopy, Singular value decomposition, Classification.
Abstract:
UV-VIS spectroscopy is a powerfull qualitative and quantitative technique used in analytical chemistry, which
gives information about electronic transitions of electrons in molecular orbitals. As in UV-VIS spectra there is
no direct information on characteristic organic groups, vibrational spectroscopy (e.g. infrared) has been pre-
ferred for biological applications. In this research, we try to use state-of-the-art fiber optics probes to obtain
UV-VIS-SWNIR diffusive reflectance measurements of yeasts and bacteria colonies on plate count agar in the
region of 200-1200nm; in order to discriminate the following microorganisms: i) yeasts: Saccharomyces cere-
visiae, Saccharomyces bayanus, Candida albicans, Yarrowia lipolytica; and ii) bacteria: Micrococcus luteus,
Pseudomonas fluorescens, Escherichia coli, Bacillus cereus. Spectroscopy results show that UV-VIS-SWNIR
has great potential for identifying microorganisms on plate count agar. Scattering artifacts of both colonies
and plate count agar can be significantly removed using a robust mean scattering algorithm, allowing also
better discriminations between the scores obtained by singular value decomposition. Hierarchical clustering
analysis of UV-VIS and VIS-SWNIR decomposed spectral scores lead to the conclusion that the use of VIS-
SWNIR light source produces higher discrimination ratios for all the studied microorganisms, presenting great
potential for developing biotechnology applications.
1 INTRODUCTION
Spectroscopy is a powerful tool for biological appli-
cations, being applicable to liquids, solutions, pastes,
powders, films, fibres, gases and surfaces, and mak-
ing possible to characterize proteins, peptides, lipids,
membranes, carbohydrates in pharmaceuticals, foods,
plants or animal tissues (Hammes, 2005).
One of the most popular method is Infrared Spec-
troscopy (IR), and was firstly applied to biological
materials in 1911 (Riddle et al., 1956). In the 1950s
and 1960s research IR spectroscopy began to be ap-
plied for microorganism differentiation, but this re-
search was abandoned due to the unsatisfactory re-
sults obtained with dispersive spectrometers (Dziuba
et al., 2007). These were ignored during 20 years,
until modern interferometric Fourier Transform Infra-
Red spectrometers (FT-IR) and statistical comput-
ing methodologies became available (Dziuba et al.,
2007).
Recent techniques using FT-IR allowed microbio-
logical characterization and the discrimination at level
of sorting better species and strains. Attenuated to-
tal reflection and IR micro-spectroscopy have been
associated to the discrimination and identification of
strains according to taxonomic classification, gram
+/- factor, or even susceptibility to antibiotics and
grown medium (Mariey et al., 2001).
FT-IR has also been used to identify lactic acid
bacteria strains (e.g. Lactobacillus, Lactococcus,
Leuconostoc, Pediococcus and Streptococcus(Dziuba
et al., 2007), and the rapid identification of Acineto-
bacter species (Winder et al., 2004). An extensive
FT-IR spectroscopy database for the identification
of bacteria from the two suborders Micrococcineae
and Corynebacterineae (Actinomycetales, Actinobac-
teria) as well as other morphologically similar gen-
era was established in 2002 by Helene Oberreuter and
its team (Oberreuter et al., 2002). Furthemore, FT-
IR was used for the first time to determine the ra-
tios of different yeast species (Saccharomyces cere-
visiae, Hanseniaspora uvarum) and two yoghurt lac-
tic acid bacteria (Lactobacillus acidophilus, Strepto-
coccus salivarius ssp. thermophilus) in suspensions
25
S. Silva J., C. Martins R., A. Vicente A. and A. Teixeira J. (2008).
FEASABILITY OF YEAST AND BACTERIA IDENTIFICATION USING UV-VIS-SWNIR DIFUSIVE REFLECTANCE SPECTROSCOPY.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 25-32
DOI: 10.5220/0001062800250032
Copyright
c
SciTePress

Table 1: Studied microorganisms characteristics and experimental conditions: Gram factor, colony colour and shape and
integration time.
Integration Time (ms)
Microorganism Gram Colony color Shape Medium UV-VIS (ms) VIS-NIR (ms)
Saccharomyces cerevisiae na white spherical YPD 70 66
Saccharomyces bayanus na white spherical YPD 45 61
Candida albicans na white spherical YPD 70 66
Yarrowia lipolytica na white rod YPD 70 61
Micrococcus luteus - yellow spherical TSA 20 19
Pseudomonas fluorescens - translucent rod MP 20 30
Escherichia coli - translucent rod LB 30 13
Bacillus cereus + opaque rod LB 36 61
of distilled water (Oberreuter et al., 2000).
Raman Spectroscopy has also shown great poten-
tial for microorganisms identifications in microscopy,
such as for Candida yeast strains and bacterial strains
such as Staphylococus, Enterococus and Echerichia
Coli) (Guibeta et al., ). Applications are also found
in oral hygiene, for the identification of Streptococ-
cus mutants, S. sanguis, S.intermedius and S. oralis
(Berger and Zhu, 2003). Moreover, this technique
is currently used to identify baterials cells of Staphy-
lococcus under different cultivation conditions (Harz
et al., 2005) and single yeast cells (Rch et al., 2005).
Fluorescence Spectroscopy (FS) is one of the most
important spectroscopic techniques in molecular bi-
ology, and consequently can also be used to microbi-
ological identification. FS applications can be found
on the differentiation of yeast and bacteria, by their
intrinsic fluorescence to UV excitation (Bhatta et al.,
2005).
UV-VIS-SWNIR spectroscopy is one of the most
widely used techniques in analytical chemistry, but it
has almost not been used for microorganism identi-
fication. This is perhaps attributed to the fact that
UV-VIS spectroscopy records transmitions between
electron energy levels from molecular orbitals, in-
stead of vibrational or structural oscillation of molec-
ular groups as in the infrared region. It is widely ac-
cepted that vibrational spectroscopy is more adequate
for organic chemistry measurement than transitional
spectroscopy. Nevertheless an asset of this technique
has never been done use for microbiological identifi-
cation.
Electronic transitions in the UV-VIS region de-
pend upon the energy involved. For any molecular
bound (sharing a pair of electrons), orbitals are a mix-
ture of two contributing orbitals σ and π, with cor-
responding anti-bounding orbitals σ
∗
and π
∗
, respec-
tively. Some chemical bounds present characteristic
orbital conditions, ordered by higher to lower order
energy transitions: i) alkanes (σ → σ
∗
; 150nm); ii)
carbonyls (σ → π
∗
; 170nm); iii) unsaturated com-
pounds (π → π
∗
; 180nm); iv) molecular bounds to
O, N, S and halogens (n → σ
∗
; 190nm); and v) car-
bonyls (n → π
∗
; 300nm). As most UV-VIS spec-
trometers yield a minimum wavelength of 200nm, this
technique has been considered to provide lower infor-
mation in terms of functional groups when compared
to IR, being the spectral differences mostly attributed
to conjugated π → π
∗
transitions and n → π
∗
tran-
sitions (Perkauparus et al., 1994).
Only recording π → π
∗
and n → π
∗
tran-
sitions above the 200nm is however not totally a
handicap. Many organic molecules present con-
jugated unsaturated and carbonyls bounds, such as
aminoacids, phospholipids, free fatty acids, phe-
nols and flavonoids, peroxides, peptides and pro-
teins, sugars and their polymers absorbance in these
bands. Furthermore, many biological molecules
present chromophore groups, which increase the ab-
sortion in the UV-VIS region, such as: nitro, nitroso,
azo, azo-amino, azoxy, carbonyl and thiocarbonyl,
which can be used to identify microorganisms.
UV-VIS-SWNIR has some advantages to FTIR
for microbiological identification in plate count agar.
The lower wavelength turns this radiation attractive
due to the lower penetration, being easier to mon-
itor surfaces than NIR or MIR radiation. Further-
more, state-of-the-art fiber-optics miniature UV-VIS-
SWNIR are today affordable for mobile applications
such as identification of microorganisms in surfaces,
using spectroscopy may became feasible in a near
future. Although UV-VIS retrieves only molecular
spectroscopy information, today’s equipments also
include high frequency vibrational spectroscopy in
the SWNIR region, giving important information on
water, fats and proteins which may be used to discrim-
inate between microorganisms.
In this research was tried to discriminate both
Yeasts and Bacteria of comonly used in microbi-
ology laboratories: i) yeasts: Saccharomyces cere-
visiae, Saccharomyces bayanus, Candida albicans,
Yarrowia lipolytica, ii) bacteria: Micrococcus luteus,
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
26

Figure 1: Yeast and bacteria growth media: (a) Tryptic Soy
Agar (TSA): Micrococcus luteus; (b) Pseudomonas Iso-
lation Agar (MP) (Pseudomonas fluorescens); (c) Luria-
Bertani (LB): Escherichia coli, (d) LB: Bacillus cereus;
(e) YPD (Saccharomyces cerevisiae); (f) YPD (Saccha-
romyces bayanus); (g) YPD (Candida albicans); and (h)
YPD (Yarrowia lipolytica).
Pseudomonas fluorescens, Escherichia coli, Bacillus
cereus; under plate count agar growth media.
The physical properties of the UV-VIS-SWNIR
spectra can also provide a great potential for the iden-
tification of microorganism, using multivariate statis-
tical analysis and signal processing techniques. Mi-
crobes may not be directly identified by their main
colony chemical composition but rather by charac-
teristic metabolites produced under different growth
media. This is especially true for yeasts that ex-
hibit one of the most complex metabolisms in this
study. Therefore, not only the colony but changes
in the composition of the plate count agar in the sur-
roundings of each colony are expected to affect the
UV-VIS-SWNIR spectra in order to obtain significant
discrimination between the different microorganisms
spectra. Therefore, the main objective of this research
work were to investigate the discrimination potential
of UV-VIS and VIS-NIR wavelengths to classify the
following microorganisms: i) yeasts: Saccharomyces
cerevisiae, Saccharomyces bayanus, Candida albi-
cans, Yarrowia lipolytica, ii) bacteria: Micrococcus
luteus, Pseudomonas fluorescens, Escherichia coli,
Bacillus cereus; under plate count agar growth media.
2 MATERIALS AND METHODS
2.1 Sample Preparation
The microorganisms were obtained from the mi-
crobiological collection of the IBB - Institute for
Biotechnology and Bioengineering at the University
of Minho. The microorganisms were incubated un-
der aerobic conditions at 35
o
C during 72h. Micrococ-
cus luteus was cultivated on Difco Tryptic Soy Agar
(TSA) while Pseudomonas fluorescens was grown on
Difco Pseudomonas Isolation Agar (MP). Escherichia
coli and Bacillus cereus were cultivated on Difco
Luria-Bertani Agar (LB). Yeast strains were grown on
Difco YPD Agar (YPD), at the same temperature and
time (Difco, 2005).
2.2 Spectroscopy
Yeast and bacteria UV-VIS-SWNIR spectroscopy
analysis was performed using the fiber optic spec-
trometer AvaSpec-2048-4-DT (2048 pixel, 200-
1100nm). Standart reflection UV-VIS and VIS-
SWNIR probes, models FCR-7UV200-2ME and
FCR-7IR200-2-ME (Avantes, 2007). A xenon and
halogen light sources, models AvaLight XE-2000
and AvaLight-Hal were used for UV-VIS and VIR-
SWNIR transmission measurements respectively; and
recorded using AvaSoft 6.0 (Avantes, 2007). Trans-
mission measurements were performed at the room
temperature of 18±2
o
C, and: (a) UV-VIS: the xenon
lamp was let to stabilize during 20 min; (b) VIS-NIR:
the tungsten lamp lamp was let to stabilize during 15
min. The dark spectra was recorded and measure-
ments were taken with linear and electric dark cor-
rection. Both light spectra were monitored by statisti-
cally assessing the reproducibility of the light source
with measurements of light during the several days
of the experiment. Fifteen spectra replicates were
recorded of UV-VIS and VIS-SWNIR measurement
of both plate count agar and microorganisms colonies
to study scattering effects. Futhermore, spectra were
obtained inside a box designed to isolate the environ-
mental light and maintain the probe at 90
o
angle with
the plate agar.
2.3 Spectral Analysis
2.3.1 Spectra Pre-processing
Table 1 presents the UV-VIS and VIS-NIR spectra ac-
quisition conditions. Experimental setup has shown
that it is impossible to use the same integration time
for the different microorganisms. Under these cir-
cumstances, all the collected spectra were normalized
x
norm
to remove this effect:
x
norm
i
= D
S
×
x
raw
i
max(x
i
)
(1)
where x
raw
i
is the original spectra, D
S
is the detec-
tor saturation value (14000 counts) and x
norm
i
the i’th
spectra normalized by its maximum value and resized
to the detector saturation.
Furthermore, as most plaque count agar growth
media are translucid, the signal recorded is in major-
ity the media information. To increase spectral vari-
FEASABILITY OF YEAST AND BACTERIA IDENTIFICATION USING UV-VIS-SWNIR DIFUSIVE
REFLECTANCE SPECTROSCOPY
27

400 500 600 700 800 900 1000
4 5 6 7 8 9
(a)
Wavelength (nm)
log(I)
400 500 600 700 800 900 1000
5 6 7 8 9
(b)
Wavelength (nm)
log(I)
400 600 800 1000
3 4 5 6 7 8 9
(c)
Wavelength (nm)
log(I)
400 500 600 700 800 900 1000
4 5 6 7 8 9
(d)
Wavelength (nm)
log(I)
Figure 2: Plaque count agar spectra: (a) raw UV-VIS; (b)
MSC UV-VIS; (c) raw VIS-NIR; (d) MSC VIS-NIR.
ance, the normalized media spectra matrix was sub-
tracted to the microorganisms spectra, obtaining the
spectra matrix (x), which is thereafter subjected to
robust mean scattering correction, and singular value
decomposition.
2.3.2 Robust Mean Scattering Correction
The collected spectra were smoothed by using a
Savisky-Golay filter (length = 4, Order= 2) (?) prior
to any exploratory data analysis procedure. After-
wards, the spectra was pre-processedusing a modified
multiplicative scatter correction algorithm (Gallagher
et al., 2005; Martens and Stark, 1991; Martens et al.,
2003). Each spectra is corrected by using the follow-
ing equation:
x
corr
= xb+ a = x
ref
(2)
The a and b are computed by minimizing the fol-
lowing error:
e
j
= bx
j
+ a− x
ref
(3)
where the x
j
is the j sample spectra and x
ref
is a
reference spectra.
This RMSC algorithm is based on the application
of the robust least squares method to determine the a
and b matrices ensuring that spectral areas that do not
correspond to scattering artifacts are not taken into
account. The robust least squares algorithm is im-
plemented by the re-weighted least squares with the
weights computed by using the Huber function. The
algorithm high breakdown point (50%) means that ex-
istent outliers will not distort the model fitting (eq. 3)
and thus, the a and b scatter correction parameters are
determined using only consistent spectral areas. The
iterative algorithm can be described, briefly as follow:
1) set the reference spectra (x
ref
) equal to the sample
−10 −5 0 5
−1.0 −0.5 0.0 0.5 1.0
PC1(96.40%)
PC2(2.03%)
(a)
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
−4 −2 0 2 4 6
−0.5 0.0 0.5
PC1(98.60%)
PC2(0.7%)
(b)
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
LB
MP
MP
MP
MP
MP
MP
MP
MP
MP
MP
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
TSA
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
YP
Figure 3: Growth media Gabriel plot: (a) UV-VIS (PC1
(96.40%), PC2 (2.3%) and (b) VIS-NIR (PC1 (98.60%),
PC2 (0.70%) to the corresponding growth media: TSA (),
MP (◦), LB (•) and YPD (⋄).
spectra closest to the median spectra; 2) correct the
remaining sample spectra by applying the above de-
scribed robust least squares procedure; eq.3) recom-
pute the median spectra and iterate until convergence.
2.3.3 Singular Value Decompostion
Singular value decomposition (SVD) is a blind signal
technique widely used in spectroscopy data, where
the corrected spectra (x
corr
) is decomposed in order of
magnitude of variation directions in the variable space
(wavelengths). Generally, most variability is captured
in the first principal components (PC), where as, in
good signal to noise spectral data, noise is captured
in the last orthogonal decompositions, and therefore a
spectra can be decomposed as:
x
corr
=
b
x+ ε(x) (4)
where the
b
x is the signal and ε(x) is the estimated
noise of x. Spectra matrix x
corr
can be decomposed
by (SVD), where:
x = USV
T
(5)
where US are the scores, V
T
the loadings and
the S singular values, respectively (Jolliffe, 1986;
Krzanowski, 1998; Baig and Rehman, 2006).
To distinguish between the number of relevant de-
compositions, one can determine the number of rele-
vant singular values by performing n randomizations
of the original spectra matrix (x) (Manly, 1998). In
this research, 5000 randomizations were performed
by rotating the spectral scope value at the same wave-
lengths among the different samples, in order to not
violate the spectral continuity. Singular values from
the original spectra x above the 1st singular value of
randomized spectra (x
rand
) define the number of inde-
pendent singular values of the original signal where is
possible to discriminate the different microorganisms
spectra:
b
x = US
relv
V
T
relv
(6)
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
28

400 500 600 700 800 900 1000
5 6 7 8 9
(a)
Wavelength (nm)
log(I)
400 500 600 700 800 900 1000
5 6 7 8 9
(b)
Wavelength (nm)
log(I)
400 600 800 1000
3 4 5 6 7 8 9
(c)
Wavelength (nm)
Log(I)
400 500 600 700 800 900 1000
3 4 5 6 7 8 9
(d)
Wavelength (nm)
log(I)
Figure 4: Microorganisms spectra: (a) raw UV-VIS; (b)
MSC UV-VIS; (c) raw VIS-NIR; (d) MSC VIS-NIR.
Where US
relv
and V
T
relv
are the statistically rele-
vant scores and loading of x, respectively. To further
discriminate between the microorganisms spectra, the
relevant PC’s scores(US
relv
) were subjected to hierar-
chical clustering according to the euclidean distance,
to determine the potential of using UV-VIS and VIS-
NIR to identify the studied yeast and bacteria. All
statistical computing analysis were performed using
R (R-Project, 2006).
3 RESULTS AND DISCUSSION
3.1 Spectral Analysis
Figure 2 presents the UV-VIS and VIS-NIR plate
count agar growth media spectra, respectively. It is
possible to observe in Figure 2(a) and 2(c) that all
plate count agar are highly dispersive, generating a
high scattering effect. This undesired scattering ef-
fect is due to the light path length to be very sensitive
to the probe angle, particles in the agar, surface tex-
ture of both agar and petri disk. If the light scattering
effect is not corrected, variance due to this physical
phenomenon affects significantly the chemical inter-
pretation of the spectra due to scattering artifacts.
As pure additive effects of light scattering are
rarely observed in samples with complex compo-
sitions, being mainly of multiplicative origin, the
growth media spectra was subjected to RMSC, be-
ing the corrected spectra presented in Figures 2 (b)
and 2 (c), respectively. By directly comparing Fig-
ures 2 (a)-(b), and 2 (c)-(d), one can observe the scat-
tering effect is obtained in both light sources. After
applying the RMSC one can observe that, this scat-
tering artifacts are significantly reduced in the region
of 700-1000nm, but nevertheless both light sources
present higher degree of spectral variance in the re-
gion of 400-700nm.
Furthermore, it is observable that all spectra are
proportional to each other. Variation is mostly in
terms of signal intensity than in spectra shape. In this
sense it is difficult to distinguish the different growth
media by direct spectra comparison. Figure 3 presents
the Gabriel plot (PC1 vs PC2) of the growth media
spectra, for UV-VIS and VIS-NIR wavelengths, re-
spectively. Both UV-VIS and VIS-NIR biplots evi-
dence optical properties of the growth media, being
thin mostly described by the 1st principal component.
In both biplots, it is possible to observe that the MP
growth media is the most translucid and YPD the
most opaque, and therefore PC1 can be interpreted as
the amount of signal that the detector records. Media
such as TSA and LB present similar spectra records.
Such is mainly attributed to their similar composition
in terms of main componentssuch as sodium chloride,
agar and water.
Results show that MP and YPD media are more
suceptible to variability than TSA and LB in spec-
troscopy terms. As no chemical assessment was per-
formed to the media, we cannot present the cause for
this source of variation.
Figure 4 presents the UV-VIS and VIS-NIR mi-
croorganisms spectra, respectively. Similarly to the
growth media, the microbe spectra exhibits high scat-
tering artifacts (see Figures 4a and 4c). The scatter-
ing effect is in this case due to the light scattering at
the colony surface and growth media, which signifi-
cantly affects the observed spectra. Similarly to the
growth media, scattering is significantly high in both
light sources and evenly distributed from 450 to 1000
nm, nevetheless scattering is significantly reduced by
the RMSC algorithm. The corrected spectra presents
higher variability in the region of 400-700nm, but
nevertheless it is difficult to recognize directly spec-
tral characteristics that distinguish the different mi-
croorganisms under study, and therefore SVD analy-
sis is necessary.
3.2 Singular Value Decomposition
Analysis
Figure 5 presents the Gabriel plot of the first
two components obtained by SVD (PC1(78.40%),
PC2(8.03%)), and the corresponding hierarchical
clustering analysis. The first two decompositions en-
sure the majority of the spectral variation to pro-
portional to the average spectrum (PC1(78.40%)),
and that linear variance also with discriminant power
(8.03%), evidences smaller but discriminate spectral
FEASABILITY OF YEAST AND BACTERIA IDENTIFICATION USING UV-VIS-SWNIR DIFUSIVE
REFLECTANCE SPECTROSCOPY
29
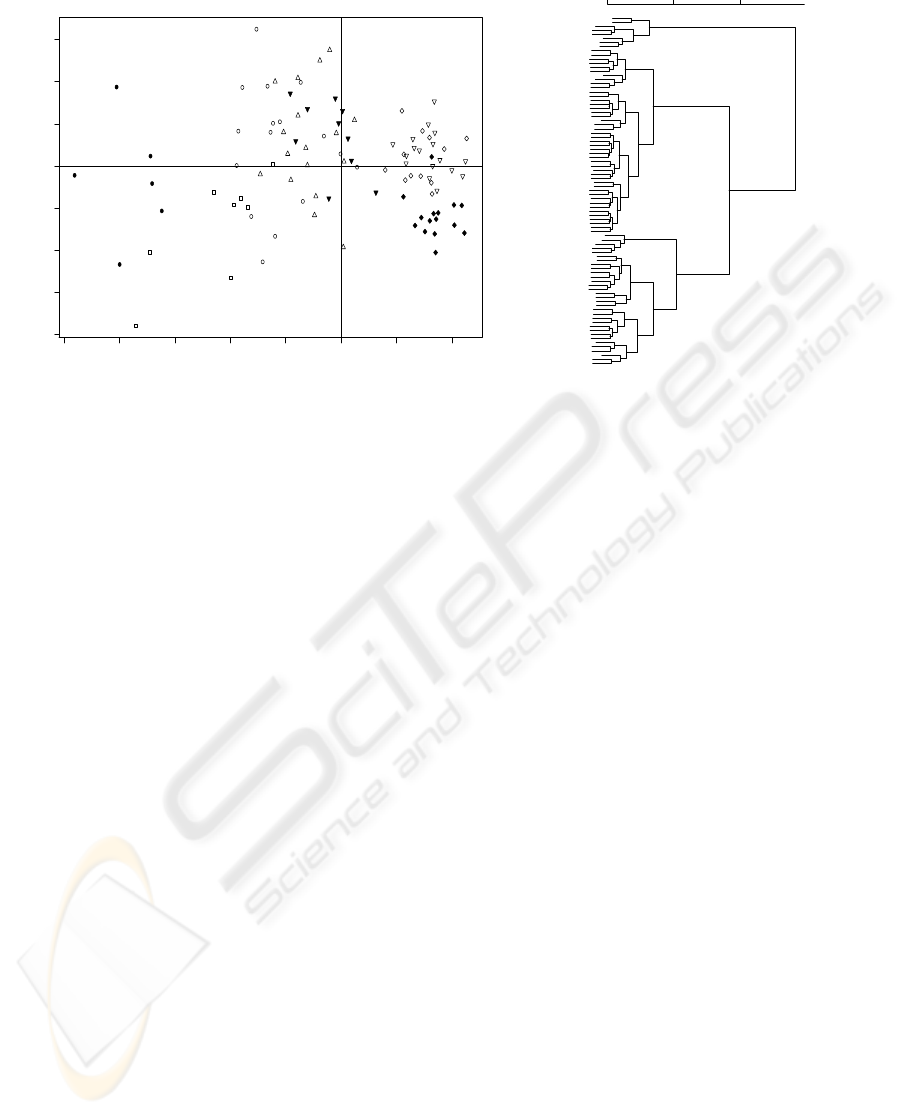
−10 −8 −6 −4 −2 0 2 4
−2.0 −1.5 −1.0 −0.5 0.0 0.5 1.0 1.5
PC1(78.40%)
PC2(8.03%)
(a)
M.Luteus
M.Luteus
M.Luteus
M.Luteus
M.Luteus
M.Luteus
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
M.luteus
M.luteus
M.luteus
M.luteus
M.luteus
M.luteus
E.coli
E.coli
C.albicans
C.albicans
B.cereus
B.cereus
B.cereus
B.cereus
S.cerevisiae
C.albicans
S.cerevisiae
C.albicans
S.cerevisiae
S.cerevisiae
B.cereus
C.albicans
C.albicans
S.cerevisiae
S.cerevisiae
Y.lipolytica
S.cerevisiae
C.albicans
S.cerevisiae
C.albicans
B.cereus
C.albicans
C.albicans
S.cerevisiae
C.albicans
S.cerevisiae
C.albicans
S.cerevisiae
C.albicans
C.albicans
B.cereus
B.cereus
B.cereus
C.albicans
C.albicans
S.cerevisiae
C.albicans
B.cereus
B.cereus
B.cereus
B.cereus
B.cereus
B.cereus
E.coli
E.coli
E.coli
E.coli
E.coli
S.bayanus
S.bayanus
S.bayanus
S.bayanus
S.bayanus
Y.lipolytica
Y.lipolytica
S.bayanus
S.bayanus
S.bayanus
Y.lipolytica
E.coli
S.bayanus
S.bayanus
S.bayanus
S.bayanus
Y.lipolytica
S.bayanus
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
S.bayanus
S.bayanus
S.bayanus
S.bayanus
Y.lipolytica
0 5 10
(b)
Height
Figure 5: UV-VIS Microorganisms Spectra PCA Analysis: (a) Gabriel Plot (PC1 (78.40%), PC2 (8.03%); S. cerevisiae (⋄), S.
bayanus (△), C. albicans (▽), Y. lipolytica (H), M. luteus (•), P. fluorescens (◦), E. coli () and B. cereus (); (b) hierarquical
clustering of Microorganisms.
differences between the studied microorganisms.
PC1(78.40%) discriminates between spectral in-
tensity, being possible to observe that colonies of
B. cereus, C. albicans and S.cerevisiae, present the
higher scores indicating that the colonies of this mi-
croorganisms are well suited for diffusive reflectance.
Although the relative proximity in the scores space, it
is observable that B. cereus, C. albicans and S. cere-
visiae are discriminated by the 2nd PC. In the group,
it is possible to observe a higher similarity between C.
albicans and S.cerevisiae spectra than with B. Cereus.
It is further observable that although S. bayanus
and Y. lipolityca exhibit similar spectral intensity,
their spectra is possible to he discriminated in the 2nd
PC. S. bayanus who presents larger variability then Y.
lipolityca, being more difficult to identify.
E. coli and M. luteus colonies are distinguish-
able from all the other microorganisms group. These
present the lowest signal intensity, and the proximity
of the two spectra may in part he due to the growth
media spectral similarity between TSA and LB, as
show in Figure 3a. Nevertheless, the UV-VIS spectra
decomposition is capable of discriminating between
E. coli and M. luteus spectra.
In the UV-VIS, P. fluorescens spectra has proved
to he highly unreproducible, being its scores
well spread throughout the 2nd PC. This un-
reproducibility is attributed to the high translucent
of both P. fluorescens colonies and the MP growth
media, and to the experimental microbiological tech-
nique. As microorganisms were inoculated using a
inoculating loop, a significant part of radiation is dif-
fused into de media in P. fluorescens. Better growth
may in the future improve the spectral measurement
and therefore its identification, as already observed in
E. coli colonies, which although are smaller, are ca-
pable of developing thicker colonies.
Dispite the experimental difficulties hierarchical
clustering analysis presented in Figure 5b shows that
the majority of the studied microorganisms cluster to-
gether with exception of P. fluorescens. This gives
good perspectives of using UV-VIS spectroscopy for
microorganisms identification in plate count agar af-
ter experimental and signal processing improvements.
The VIS-SWNIR spectra also exhibits high scat-
tering artifacts in the 400-1000nm region (see Fig-
ure 4c), the scattering effect is effectively removed
by the RMSC algorithm used during the spectral pre-
processing. Figure 4d, shows that in the corrected
spectra, variance is higher in the 400-700nm as al-
ready observed as with the UV-VIS light source. The
majority of the spectra are proportional to each other,
varying only in signal intensity.
Figure 6a presents the Gabriel plot of the first
two PCs of the decompose VIS-NIR spectra (PC1
(75.40%), PC2 (10.95%), and the corresponding
scores hierarchical clustering. Similarly to the UV-
VIS spectra, the first spectra decomposition discrim-
inating variance proportional to the average spec-
tra, and PC2 (lower variance) tends to discriminate
smaller details between the spectra of the studied mi-
croorganisms, evidencing that VIS-NIR is a viable
methodology for identification microorganisms on
plate count agar. PC1 (75.40%) of VIS-SWNIR spec-
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
30
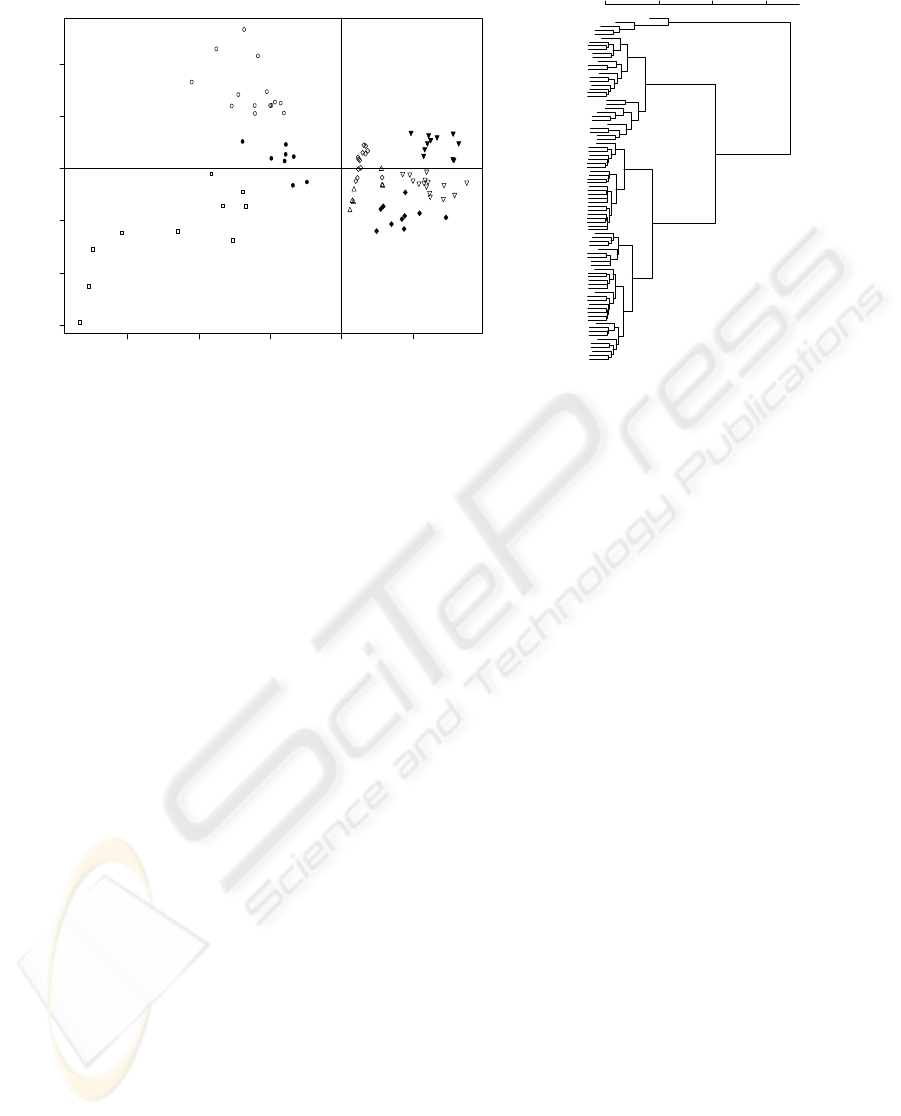
−15 −10 −5 0 5
−3 −2 −1 0 1 2
PC1(75.40%)
PC2(10.95%)
(a)
M.Luteus
M.Luteus
M.Luteus
M.Luteus
M.Luteus
M.Luteus
M.Luteus
M.Luteus
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
P.Flurescencis
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
E.Coli
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
B.Cereus
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Cerevisiae
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
S.Bayanus
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
C.Albicans
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
Y.Lipolytica
E.coli
E.coli
E.coli
E.coli
E.coli
M.luteus
M.luteus
M.luteus
M.luteus
M.luteus
M.luteus
P.fluorescens
P.fluorescens
P.fluorescens
M.luteus
P.fluorescens
P.fluorescens
P.fluorescens
P.fluorescens
P.fluorescens
P.fluorescens
P.fluorescens
P.fluorescens
P.fluorescens
M.luteus
P.fluorescens
P.fluorescens
E.coli
E.coli
E.coli
E.coli
E.coli
B.cereus
B.cereus
B.cereus
S.bayanus
S.cerevisiae
S.bayanus
S.bayanus
S.bayanus
S.bayanus
S.bayanus
S.bayanus
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
S.cerevisiae
C.albicans
B.cereus
C.albicans
C.albicans
C.albicans
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
Y.lipolytica
B.cereus
C.albicans
C.albicans
C.albicans
C.albicans
C.albicans
C.albicans
C.albicans
B.cereus
B.cereus
B.cereus
B.cereus
Y.lipolytica
B.cereus
C.albicans
C.albicans
C.albicans
C.albicans
0 5 10 15
(b)
Height
Figure 6: VIS-NIR Microorganisms SpectraPCA Analysis: (a) Gabriel Plot (PC1 (75.40%), PC2 (10.95%); S. cerevisiae.
(⋄), S. bayanus (△), C. albicans (▽), Y. lipolytica (H), M. luteus (•), P. fluorescens (◦), E. coli () and B. cereus (); (b)
hierarquical clustering of Microorganisms.
tra presents a clear discrimination between groups
of microorganisms: (a) B. cereus, S.cerevisiae, S.
bayanus, Y. lipolityca, C. albicans, and (b) M. luteus,
E. coli and P. fluorescens. PC2 (10.95%) is capa-
ble of discriminate the microorganisms inside these
two groups, and therefore, complete discrimination is
achievable with VIS-SWNIR wavelengths.
In the VIS-SWNIR spectra E. coli and P. fluo-
rescens present higher dispersion of signal when com-
pared to the rest of the spectra of microorganisms.
Nevertheless, results reproductivity inside these two
groups is significantly higher than with the UV-VIS
measurements; indicating that translucid colonies
were better identified under the VIS-SWNIR radi-
ation. Furthermore, data suggest that VIS-SWNIR
may be better to be used under non-optimal measur-
ing conditions. This is especially problematic if we
want to identify microorganisms in plate count agar
with similar compositions and metabolism.
3.3 Methodology Improvements
This preliminary study shows that UV-VIS and VIS-
SWNIR reflection spectroscopy has a great potential
for rapid qualitative discrimination of yeasts and bac-
teria in plate count agar. Nevertheless, both experi-
mental methodologyand signal processing techniques
should be improved to take advantage of the informa-
tion contained in the UV-VIS-SWNIR.
Improvements to the experimental methodology are
necessary to improve discrimination in the studied re-
gion of the spectra. For example, the use of liquid
cultures to replicate microorganisms and then innoc-
ulate by a droplet on the surface agar will allow the
growth of compact and thicker colonies. Small and
thin colonies lead to readings with much media infor-
mation, because the light is reflected from both sites
of the agar. This is particularly relevant for E. coli
and P. fluorescens because of the small size of the
colonies and small thickness of P. fluorescens; being
the spectra highly affected by the media composition,
leading to sistematic errors in discrimination. Mi-
croorganisms signal spectra can also be maximised
by reducing the growth media thickness. Such min-
imises light dispersion accross the agar, and increases
the colony spectral intensity by passing through it the
reflected light; ans as well by optimising fiber optics
diffusive reflectance position and angle control, min-
imizing scattering effects in the colonies and growth
media.
Scattering artifacts and small noise were success-
fully removed by pre-processing the spectra with the
RMSC and Savisky-Golay filter, respectively; being
possible to achieve high-quality and resolution final
spectra before signal treatment. Improvements can
also be performed to the spectra processing proce-
dure. Methods such as the combination of logis-
tic partial least squares (log-PLS) with multiblock
(e.g. UV-Vis+Vis-Nir spectra) can provide a new in-
sight to the discrimination of microorganisms using
spectroscopy. If these methodologies provide good
discriminations, more robust techniques such as the
use of wavelets for compressing the original spectra
and orthogonal-PLS classification of spectra will be
tested.
FEASABILITY OF YEAST AND BACTERIA IDENTIFICATION USING UV-VIS-SWNIR DIFUSIVE
REFLECTANCE SPECTROSCOPY
31

4 CONCLUSIONS
Results show that UV-VIS-SWNIR spectroscopy is a
feasable technology for plate count agar microorgan-
isms identifications. The robust mean scattering cor-
rection algorithm was able to efficiently remove the
growth media and colonies scattering artifacts, allow-
ing a better interpretation of the singular value de-
composition scores loading. In this exploratory ex-
periment, VIS-SWNIR wavelengths were able to pro-
duce better discriminations between microorganisms
than the UV-VIS region. Nevertheless, experimen-
tal methodology and signal processing improvements
proposed may increase the discrimination resolution,
making UV-VIS-SWNIR an attractive methodology
for rapid microorganisms identification in plate count
agar.
REFERENCES
Avantes (2007). Avaspec-2048-4-dt/-rm.
http://www.avantes.com.
Baig, S. and Rehman, F. (2006). Signal modeling using sin-
gular value decomposition. In Advances in Computer,
Information, and Systems Sciences, and Engineering.
Springer Netherlands.
Berger, A. and Zhu, Q. (2003). Identification of oral bacte-
ria by raman microspectroscopy. In Journal of Mod-
ern Optics 50(15-17): 2375-2380. Taylor and Francis
Group.
Bhatta, H., Goldys, E., and Learmonth, R. (2005). Rapid
identification of microorganisms by intrinsicfluores-
cence. In Imaging, Manipulation, and Analysis of
Biomolecules and Cells: Fundamentals and Applica-
tions III,. SPIE.
Difco, L. (2005). Difco manual : dehydrated culture media
and reagents for microbiology. - VII. Difco Laborato-
ries, Detroit, USA.
Dziuba, B., Babuchowski, A., Naleczb, D., and Niklewicz,
M. (2007). Identification of lactic acid bacteria using
ftir spectroscopy and cluster analysis. In International
Dairy Journal 17: 183189. Elsevier.
Gallagher, N. B., Blake, T., and Gassman, P. (2005). Ap-
plication of extended inverse scattering correction to
mid-infrared reflectance of soil. In Journal of Chemo-
metrics 19: 271-281.
Guibeta, F., Marieya, L., Pichonb, P., Traverta, J., Denisb,
C., and Amiela, C. Discrimination and classifica-
tion of enterococci by fourier transform infrared spec-
troscopy.
Hammes, G. G. (2005). Fundamentals of spectroscopy. In
Spectroscopy for the Biological Sciences. John Wiley
Sons, Inc.
Harz, M., Rosch, P., Peschke, K.-D., Ronneberger, O.,
Burkhardtb, H., and Popp, J. (2005). Micro-raman
spectroscopic identification of bacterial cells of the
genus staphylococcus and dependence on their cul-
tivation conditions. In Analyst 130: 15431550. The
Royal Society of Chemistry.
Jolliffe, I. (1986). Principal Component Analysis. Springer,
New York, USA.
Krzanowski, W. J. (1998). Principles of Multivariate Data
Analysis. Oxford University Press, Oxford, UK.
Manly, B. F. (1998). Randomization, Bootstrap and Monte
Carlo Methods in Biology. Chapman and Hall, Lon-
don, UK, 2nd edition.
Mariey, L., Signolle, J., Amiel, C., and Travert, J. (2001).
Discrimination, classification, identification of mi-
croorganisms using ftir spectroscopy and chemomet-
rics. In Vibrational Spectroscopy 26: 151-159. Else-
vier.
Martens, H., Nielsen, J. P., and Engelsen, S. B. (2003).
Light scattering and light absorbance separated by ex-
tended multiplicative signal correction. application to
near-infrared transmission analysis of powder mix-
tures. In Analytical Chemistry 75(9): 394-404. Amer-
ican Chemical Society.
Martens, H. and Stark, E. (1991). Extended multiplicative
signal correction and spectral interference subtraction:
new preprocessing methods for near infrared spec-
troscopy. In Journal of Pharmaceutical and Biomedi-
cal Analysis 9: 625-635. American Chemical Society.
Oberreuter, H., Mertens, F., Seiler, H., and Scherer, S.
(2000). Quantification of micro-organisms in binary
mixed populations by fourier transform infrared (ft-
ir) spectroscopy. In Letters in Applied Microbiology
2000, 30: 8589. The Society for Applied Microbiol-
ogy.
Oberreuter, H., Seiler, H., and Scherer, S. (2002). Iden-
tification of coryneform bacteria and related taxa by
fourier-transform infrared (ft-ir) spectroscopy. In In-
ternational Journal of Systematic and Evolutionary
Microbiology 52: 91100. IUMS.
Perkauparus, H., Grinter, H., and Therfall, T. (1994). Uv-Vis
spectroscopy and its applications. Springer-Verlag,
New York, USA.
R-Project (2006). R: A programming environment for data
analysis and graphics. URL: http://www.r-project.org.
Rch, P., Harz, M., Schmitt, M., and Popp, J. (2005). Raman
spectroscopic identification of single yeast cells. In J.
Raman Spectrosc 36: 377379. Wiley InterScience.
Riddle, J., Kabler, P., Kenner, B., Bordner, R., Rockwood,
S., and Stevenson, H. (1956). Bacterial Identification
by Infrared Spectrophotometry. Cincinnati, Ohio.
Winder, C., Carr, E., Goodacre, R., and Seviour, R. (2004).
The rapid identification of acinetobacter species using
fourier transform infrared spectroscopy. In Journal
of Applied Microbiology 96, 328339. The Society for
Applied Microbiology.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
32
