
MANAGING MEDICINAL INSTRUCTIONS
Juha Puustjärvi
Helsinki University of Technology, Innopoli 2, Tekniikantie 14, Espoo, Finland
Leena Puustjärvi
The Pharmacy of Kaivopuisto, Neitsytpolku 10, Helsinki, Finland
Keywords: Medicinal documents, Information retrieval, Taxonomies, Ontologies, Business process management.
Abstract: The number of new medications increases every year. As a result also the amount of new instructions
concerning new medication increases rapidly. A problem is how to ensure that the employers of the
medicinal organizations are aware of the relevant medicinal instructions. In this paper, we restrict ourselves
on this problem. In particular, we consider three complementary ways for the dissemination of medicinal
instructions: (i) by providing keyword-based searching of instructions (ii), by providing ontology-based
searching of instructions, and (iii) by integrating the instructions to employers´ day-to-day work tasks. Our
argument is that integration is most preferable as medicinal instructions are provided just-in-time, tailored to
their specific needs, and integrated into day-to-day work patterns. However, automating the integration of
instructions to day-to-day work pattern is not an easy task. In our solution, day-to-day work patterns are
described by BPMN (Business Process Modeling Notation) and BPMN’s association- notation is used for
integrating the instructions to BPMN-processes. The integration of the tasks and instructions is based either
on a medicinal ontology or a taxonomy. The ontology specifies the relationships of the day-to-day tasks and
the medicinal instructions. The taxonomy is used for attaching metadata items for the tasks and instructions,
and so the integration of the tasks and instructions can be done based on the similarity of their metadata
descriptions.
1 INTRODUCTION
Healthcare is a field where the fast development of
drug treatment and technologies requires specialized
skills and knowledge that need to be renewed
frequently (Puustjärvi and Puustjärvi, 2006).
Furthermore, the number of new medications
increases every year. As each drug has its unique
indications, cross-reactivity, complications and costs
also the prescribing medication as well as the
distribution of medicinal products becomes still
more complex (Jung, 2005). As a result also the
amount of new instructions concerning new
medication increases rapidly. An interesting
question arising from this reality is how medicinal
instructions should be organized and retrieved in
order to ensure that the employees are aware of the
relevant medicinal instructions.
In principle, the (medicinal) information
retrieval system should be able to retrieve all the
medicinal instructions, which are relevant while
retrieving as few non-relevant instructions as
possible. This kind of quality of information
retrieval system is usually measured by two
fractions, called recall and precision (Baeza-Yates
and Ribeiro-Neto, 1999). Recall is the fraction of the
relevant documents (e.g., medicinal instructions),
which has been retrieved. Precision is the fraction of
the retrieved documents, which is relevant. The
values of these fractions are highly dependent on the
way the query and the content of medicinal
documents are presented.
In this article, we will first illustrate that by
replacing taxonomy–based searching by ontology-
based searching we can significantly improve both
the recall and precision fractions in searching
medicinal instructions.
On the other, minimizing the extra time required
for retrieving the instructions is turned out to be
crucial. Therefore, in many cases integrating
medicinal instructions to day-to-day work patterns is
105
Puustjärvi J. and Puustjärvi L. (2009).
MANAGING MEDICINAL INSTRUCTIONS.
In Proceedings of the International Conference on Health Informatics, pages 105-110
DOI: 10.5220/0001122401050110
Copyright
c
SciTePress

more preferable. Moreover, in this approach,
medicinal instructions are provided just-in-time, and
tailored to their specific needs.
However, automating the integration of
instructions to day-to-day work pattern is not an
easy task. As we will show, in our solution day-to-
day work patterns are described by BPMN (Business
Process Modeling Notation) (White, 2006) and
BPMN’s association- notation is used for integrating
the instructions to BPMN-processes. The integration
of the tasks and instructions is based either on a
medicinal ontology or a taxonomy. The ontology
specifies the relationships of the day-to-day tasks
and the medicinal instructions. The taxonomy is
used for attaching metadata items for the tasks and
instructions, and so the integration of the tasks and
instructions can be done based on the similarity of
their metadata descriptions.
The rest of the paper is organized as follows.
First, in Section 2, we give a motivating example of
the restrictions that we will encounter in using
keyword-based search in retrieving medicinal
instructions. Then, in Section 3, we illustrate the use
of medicinal ontologies in retrieving medicinal
instructions. How such ontologies can be specified
by the Web Ontology Language (OWL) is illustrated
in Section 4. Then, in Section 5, we illustrate how
day-to-day work patterns can be modeled by
business process modeling language BPMN. In
particular, we present how the modeling primitives
of BPMN can be used in attaching medicinal
instructions to business process tasks which model
the day-to-day work patterns. Finally, Section 6
concludes the paper by discussing the advantages
and disadvantages of our approach.
2 TAXONOMY-BASED
SEARCHING
Documents’ content is traditionally represented
through keywords, which are extracted directly from
the document (Baeza-Yates and Ribeiro-Neto,
1999). However, a reason for missing many relevant
documents is that the keywords used with queries
and documents descriptions are not standardized
(Puustjärvi and Pöyry, 2006). In order to standardize
semantic metadata specific taxonomies are
introduced in many disciplines. To illustrate this, a
simple drug taxonomy is presented in Figure 1. The
idea behind this classification is that the medicinal
instructions can be annotated by the metadata items
(the branching points and the leaves) represented in
the tree.
A user can then query medicinal instructions by
Boolean expressions (Baeza-Yates and Ribeiro-
Neto, 1999) comprising of operands and operations.
The operands are the used keywords (which are
taken from the taxonomy) and the operands are
typically “and”, “or”, and “not”. For example, by
using the taxonomy of Figure 1 the keywords
attached to the medicinal instruction “New warnings
of using pain drugs in topical use with children”
could be “Pain drugs for topical use” and
“Prescription based pain drug”.
Medical product category
Cough drug
Pain drug Fewer drug
Prescription
based pain
drug
Oral pain
drug
Pain drug
for topical
use
Injection
pain drug
Figure 1: Medicinal product categories in a taxomomy.
Now assume that a pharmacist has to check the
instructions concerning pain drugs, and so she enters
the Boolean expression: Prescription based pain
drug and Pain drug for topical use. In our example
the result includes at least the instructions “New
warnings of using pain drugs in topical use with
children”. After reading the instruction the
pharmacist is interested to read the previous
medicinal instruction of the same topic. The
pharmacist may also be interested to know the
medicinal products that are under this new warning.
Unfortunately by using keyword based searching
(i.e., Boolean expressions) the pharmacist has no
hope for finding the answers for such queries.
In the next section we will consider an ontology-
based (Gruber, 1993; Antoniou and Harmelen, 2004)
searching that supports such queries as well as the
queries based on taxonomies.
3 ONTOLOGY-BASED
SEARCHING
In order that the information retrieval system could
answer for the queries presented in previous section
we have to extend the search functionalities by
querying features. This requires the deployment of
an ontology.
HEALTHINF 2009 - International Conference on Health Informatics
106
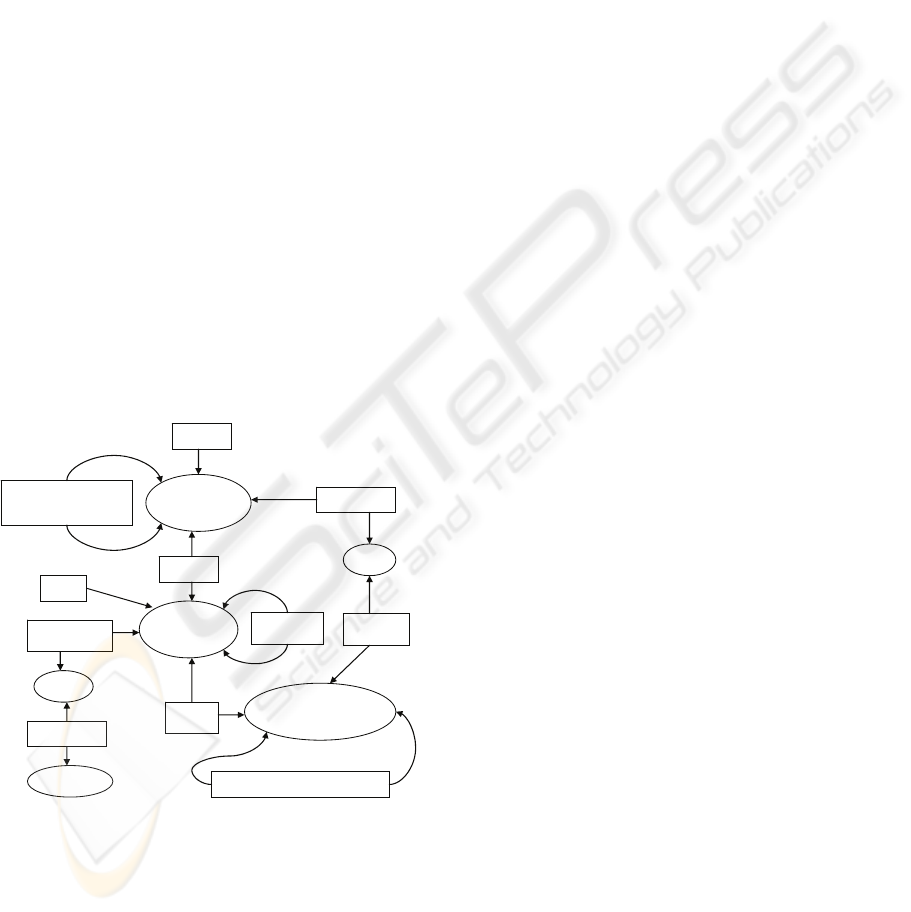
Our developed medicinal ontology models the
medicinal instructions as well as their relationships
to other relevant medicinal concepts such as patient,
physician, patient record, drug, and e-prescriptions.
In addition the ontology models the associations of
medicinal instructions to the tasks of the day-to-day
work tasks. Part of this ontology is graphically
presented in Figure 2. In the figure ellipses represent
classes and boxes represent properties. The ontology
includes for example the following information:
• Medicinal product category is a class and
each instance of the category may have a
parent, which is also an instance of
medicinal product category, i.e., among
other things, the ontology models the
taxonomy presented in Figure 1.
• Each medicinal instruction relates to zero or
more medicinal products (e.g., Aspirin),
and each medicinal product includes one or
more drugs and has one or more
substitutable medicinal products.
• Each medicinal instruction is associated to
zero or more tasks which are parts of a
workflow. That is, each medicinal
instruction is associated to a task and a
workflow which represents functionalities
in a day-to-day work patterns.
medicinal
instruction
replaces
deals
medicinal
product
includes
drug
substitutable
medicinal product
medicinal
product catrgory
deals
parent product category
task
associates
is_part_of
workflow
belongs
price
title
Figure 2: A medicinal ontology.
The ontology of Figure 2 allows making queries
such as:
• Give me medicinal instructions having
keywords Prescription based pain drug and
Pain drug for topical use (i.e., a query that
corresponds to a Boolean expression).
• Give me the medicinal instruction that is
replaced by the medicinal instruction “New
warnings of using pain drugs in topical use
with children”.
• Give me the names of the medicinal
products that relate to the medicinal
instruction “New warnings of using pain
drugs in topical use with children”.
• Assuming that the result includes the
medicinal products A and B, then it allows
querying (browsing by clicking the edges)
the substitutable medicinal products and
prices for A and B, as well as the drugs that
are included to these medicinal products.
In the next section we illustrate how our used
medicinal ontology can be specified by an ontology
language.
4 USING OWL FOR
PRESENTING MEDICINAL
INFORMATION
Web Ontology Language (OWL) (Daconta, Obrst
and Smith, 2003; OWL, 2006) has more facilities for
expressing meaning and semantics than XML, RDF
and RDF Schema, and thus OWL goes beyond these
languages in its ability to represent machine
interpretable content of the ontology (Mattocks,
2005). In particular, it adds more semantics for
describing properties and classes, for example
relations between classes, cardinality of
relationships, and equality of classes and instances.
The instances in OWL-ontologies are presented
by RDF-descriptions. RDF (Resource Description
Framework) (Davies, Fensel and Harmelen, 2002) is
essentially a data model. There are various ways in
capturing knowledge with RDF, e.g., as natural
language sentence, in a simple triple notation called
N3, in RDF/XML serialization format, and by as a
graph of the triples (Daconta, Obrst and Smith,
2003).
RDF’s modeling primitive is an object-attribute-
value triple, which is called a statement. For
example, the medicinal instruction titled “New
warnings of using pain drugs in topical use with
children” deals the medicinal products “Pain drugs
for topical use” and “Pain drugs for topical use” is
a natural language sentence that can be presented by
RDF/XML serialization format (Figure 3) by using
the vocabulary (ontology) presented in Figure 2.
However note that in capturing knowledge the
MANAGING MEDICINAL INSTRUCTIONS
107

designers are not burdened by using RDF/XML
serialization format (Antoniou and Harmelen, 2004)
as there are graphical editors that are used in design
and which automatically produce the descriptions in
RDF/XML and OWL-format (e.g., the Protégé
editor (Protégé, 2007)).
<rdf:RDF
xmlns : rdf=”http://www.w3.org/1999/02/22-rdf-syntax-ns#”
xmlns : xsd=”http://www.w3.org/2001/XMLSchema#”
xmlns : mo=“http://www.lut.fi/ontologies/medicinal_ontology#”>
<rdf:Description rdf:about=” # Instruction123”>
<rdf:type rdf:resource=“&mo;medicinal_instruction”/>
<mo : title> New warnings of using pain drugs
in topical use with children </mo : title>
<mo : deals>Pain drugs for topical use</mo : deals>
<mo : deals> Prescription based pain drugs</mo : deals>
<mo : replaces> rdf: resource
Instruction122</mo : replaces>
<mo : assocoates> rdf: resource
Check_the_dose</mo : replaces>
</rdf : Description>
</rdf:RDF
Figure 3: An RDF-statement in a medical ontology.
Note that the above illustrative RDF-description
uses the vocabulary (named medicinal_ontology)
having url
http://www.lut.fi/ontologies/medicinal_ontology#.
The rdf-description also states that the new
instruction identified by Instruction123 replace the
instruction identified by Instruction122. In addition
the description associates the instruction to a task
named “Check_the_dose”. How this association is
presented in our used business process specification
language is the topic of the next section.
5 ATTACHING INSTRUCTIONS
TO D AY-TO -D AY WO RK
PATTERNS
Though the ultimate goal of using Business Process
Modeling Notation (BPMN) (White, 2006; BPMN,
2005) is the automation of the coordination of
business processes we use BPMN to model
medicinal processes and for attaching medicinal
instructions to day-to-day work patterns which are
presented in BPMN.
The BPMN is a standard for modeling business
process flows and web services. Basically BPMN
and the UML 2.0 Activity Diagram from the OMG
(White, 2006) are rather similar in their presentation.
However, the Activity diagram has not adequate
graphical presentation of parallel and interleaved
processes, which are typical in workflow
specifications.
The BPMN defines a Business Process Diagram
(BPD), which is based on a flowcharting technique
tailored for creating graphical models of business
process operations. These elements enable the easy
development of simple diagrams that will look
familiar to most analysts. In addition BPMN allows
an easy way to connect documents and other
artifacts to flow objects, and so narrows the gap
between process models and conceptual models.
Also, a notable gain of BPMN specification is that it
can be used for generating executable BPEL
(Business Process Execution Language) (BPEL,
2004) code.
We now give an overview of the BPMN. We
first shortly describe the types of graphical objects
that comprise the notation, and then we show how
they work together as part of a Business Process
Diagram (BPD) (White, 2006). After it, we give a
simplified pharmaceutical process description using
BPD.
In BPD there are tree Flow Objects: Event,
Activity and Gateway:
• An Event is represented by a circle and it
represents something that happens during
the business process, and usually has a
cause or impact.
• An Activity is represented by a rounded
corner rectangle and it is a generic term for
a task that is performed in companies. The
types of tasks are Task and Sub-Process.
So, activities can be presented as
hierarchical structures.
• A Gateway is represented by a diamond
shape, and it is used for controlling the
divergence and convergence of sequence
flow.
In BPD there are also three kind of connecting
objects: Sequence Flow, Message Flow and
Association.
• A Sequence Flow is represented by a solid
line with a solid arrowhead.
• A Message Flow is represented by a dashed
line with an open arrowhead and it is used
to show the flow of messages between two
separate process participants.
• An Association is represented by a dotted
line with a line arrowhead, and it used to
associate data and text with flow objects.
In Figure 4, we have presented how the process
of producing electronic prescription can be
HEALTHINF 2009 - International Conference on Health Informatics
108

represented by a BPD. As illustrated in the figure we
use Association to attach instructions to Activities
and Gateways. For example, Instruction A is
associated to activity “Produce prescription”, and
Instruction B is associated to gateway “Check
negative effects”.
Produce a prescription
Check
negative
effects
Check
the dose
Check
the prices
Sign the prescription
Send the prescription to
prescription holding store
Send to expert database system
Send to medical database
Send to pricing authority
No
No
No
Yes
Yes
Yes
Instruction
A
Instruction
C
Instruction
D
Instruction
B
Instruction
E
Figure 4: Attaching medicinal instructions to a BPD.
We can support automatic integration of
medicinal instructions to business processes in two
ways. If the relationship of the medicinal instruction
and the task of a workflow (Figure 2) are presented
in the medicinal ontology, then this information can
be used.
Otherwise the integration can be made by
comparing the similarities of the metadata
descriptions of the instructions and the tasks. This
requires that the metadata items of the workflow
tasks are picked up from the same taxonomy that is
used for annotating medicinal instructions. Hence
we can conclude that an instruction is relevant for
the task, if they have similar metadata description.
That is, it is appropriate to integrate Instruction I to
Task T, if they have somehow similar keywords. To
illustrate this, assume that Instruction I has m
keywords and Task T has m keywords, then they
have at most min{m,n} common keywords. So we
can assume that the higher the number of the
common keywords is, the better the Instruction I
match for the Task T. Hence, we order the
instructions of the Task T according to the number of
their common keywords.
6 CONCLUSIONS
Healthcare is a field where the fast development of
drug treatment and technologies requires specialized
skills and knowledge. At the same time the amount
of new instructions concerning new medication
increases rapidly. How to ensure that healthcare staff
is aware of the new instructions is not an easy task.
However, applying computing technology for
retrieving and disseminating medicinal information
this complexity can be alleviated in many ways.
Particularly, we have considered taxonomy-
based and ontology-based retrieving of medicinal
instructions. It is turned out that by deploying
ontology-based retrieving method the expression
power of searching expressions can be considerably
increased. On the other hand, the drawback of
ontology-based searching is that the ontology must
be updated whenever a new medicinal instruction is
published. However, such an update can be done by
medicinal authorities, and thus it does not burden the
medicinal organizations that use the ontology.
We have also presented how the dissemination
of medicinal instructions can be carried out by
integrating the instructions to daily tasks. The gain
of this approach is that dissemination processes are
integrated in a natural way into day-to-day work
patterns, and thereby minimize the extra time
required for retrieving the instructions.
The introduction of a new technology in
retrieving and disseminating medicinal instructions
is also an investment. The investment on new ICT-
technology includes a variety of costs including
software, hardware and training costs. Introducing
and training the staff on new technology is a notable
investment, and hence many organizations like to
cut on this cost as much as possible. However, the
incorrect usage and implementation of a new
technology, due to lack of proper training, might
turn out to be more expensive in the long run.
REFERENCES
Antoniou, G., Harmelen., F., 2004. A semantic web
primer. The MIT Press.
Baeza-Yates, R., Ribeiro-Neto, B., 1999. Modern
Information Retrieval. Addison Wesley.
BPEL, 2004. Business Process Execution Language for
Web Services,
http://www.ibm.com/developerworks/library/specificat
ion/ws-bpel/
BPEL4WS, 2005. Business Process Language for Web
Sevices.
MANAGING MEDICINAL INSTRUCTIONS
109

http://www.w.ibm.com/developersworks/webservices/
library/ws-bpel/
BPMN, 2005. Business Process Modeling Notation
(BPMN), http://www.bpmn.org/
Daconta, M., Obrst, L., Smith, K., 2003. The semantic
web. Indianapolis: John Wiley & Sons.
Davies, J., Fensel, D., Harmelen, F., 2002. Towards the
semantic web: ontology driven knowledge
management. John Wiley & Sons.
Gruber, T. R., 1993. Toward principles for the design of
ontologies used for knowledge sharing. Padua
workshop on Formal Ontology.
Jung, F., 2005. XML-based prescription drug database
helps pharmacists advise their customers.
http://www.softwareag.com(xml/aplications/sanacorp.htm.
Mattocks, E. 2005. Managing Medical Ontologies using
OWL and an e-business Registry / Repository.
http://www.idealliance.org/proceedings/xml04/papers/85/
MMOOEBRR.html.
OWL, 2006. Web OntologyLanguage.
http://www.w3.org/TR/owl-ref/
Puustjärvi, J., Puustjärvi, L., 2006. The challenges of
electronic prescription systems based on semantic web
technologies. In Proc. of the 1st European Conference
on eHealth (ECEH’06). pages 251-261.
Puustjärvi, J., Pöyry, P., 2006. Information retrieval in
virtual universities. International Journal of Distance
Education Technologies (JDET) 3(4), 36-47,
July/September.
Protégé, 2007. http://protege.stanford.edu/
White, A,. 2006. Introduction to BPMN, IBM Corporation
http://www.bpmn.org/Documents/Introduction%20to
%20BPMN.pdf
HEALTHINF 2009 - International Conference on Health Informatics
110
