
COORDINATE-BASED META-ANALYTIC SEARCH
FOR THE SPM NEUROIMAGING PIPELINE
The BredeQuery Plugin for SPM5
Bartłomiej Wilkowski, Marcin Szewczyk, Peter Mondrup Rasmussen
Lars Kai Hansen and Finn
˚
Arup Nielsen
Informatics and Mathematical Modelling, Technical University of Denmark, Kongens Lyngby, Denmark
Keywords:
SPM, fMRI, PET, Neuroinformatics, Talairach, MNI, Brain region, Brain function, Brain activity, Article
retrieval, Coordinate-based search, Bibtex, Brede Database, Ontology.
Abstract:
Large amounts of neuroimaging studies are collected and have changed our view on human brain function.
By integrating multiple studies in meta-analysis a more complete picture is emerging. Brain locations are
usually reported as coordinates with reference to a specific brain atlas, thus some of the databases offer so-
called coordinate-based searching to the users (e.g. Brede, BrainMap). For such search, the publications,
which relate to the brain locations represented by the user coordinates, are retrieved. In this paper we present
BredeQuery – a plugin for the widely used SPM5 data analytic pipeline. BredeQuery offers a direct link from
SPM5 to the Brede Database coordinate-based search engine. BredeQuery is able to ‘grab’ brain location
coordinates from the SPM windows and enter them as a query for the Brede Database. Moreover, results of
the query can be displayed in an SPM window and/or exported directly to some popular bibliographic file
formats (BibTeX, Reference Manager, etc).
1 INTRODUCTION
The growing number of functional neuroimaging
studies of increasingly sophisticated human brain ac-
tivity brings the demand for new tools/services for
integration of research findings, wider exchange of
information between laboratories from the same re-
search area and efficient searching of related articles,
reviews and other literature (Wager et al., 2007).
The dominant paradigm in current neuroimaging
is that of functional localization. Functional locali-
zation hypothesizes that a given human behavior is
established by a change in brain activity in a rela-
tively limited number of spatially segregated proce-
ssing units. Thus the result of an experiment under
this paradigm consists of a Statistical Parametric Map
(SPM) indicating the local involvement. Often the
SPM is summarized as a list of regions, see e.g., (Fris-
ton et al., 2007; Pekar, 2006), in which the SPM has
been judged to be significantly different from zero
(regions were the null hypothesis is rejected). As
the typical neuroimaging experiment investigates a
highly controlled behavior and often involves a rela-
tively limited number of subjects, there is strong need
for tools to integrate multiple experiments in order to
increase the robustness to the experiment specific im-
plementation of the given behavior and to statistical
fluctuation due to limited sample sizes.
Several methods have been proposed for neu-
roimaging meta-analysis and for estimation of associ-
ations between the brain locations and textual repre-
sentations of behavior, for a recent review, see e.g.,
(Wager et al., 2007). A set of methods are based on
the so-called Brede Database (Nielsen, 2003). Me-
thods for integration include estimation of conditional
probability density functions representing the loca-
lized probability of activation in response to a given
behavior ‘word’ (Nielsen and Hansen, 2002; Nielsen
and Hansen, 2004) and multivariate methods based
on non-negative matrix factorization that aim to rep-
resent global dependencies between brain activation
and semantic text labels from neuroscience publica-
tions (Nielsen et al., 2004).
Brain locations are reported as region coordinates
relative to a specific brain atlas (usually MNI or Ta-
lairach spaces), hence, there is an interest for effec-
11
Wilkowski B., Szewczyk M., Rasmussen P., Hansen L. and Nielsen F. (2009).
COORDINATE-BASED META-ANALYTIC SEARCH FOR THE SPM NEUROIMAGING PIPELINE - The BredeQuery Plugin for SPM5.
In Proceedings of the International Conference on Health Informatics, pages 11-17
DOI: 10.5220/0001510000110017
Copyright
c
SciTePress
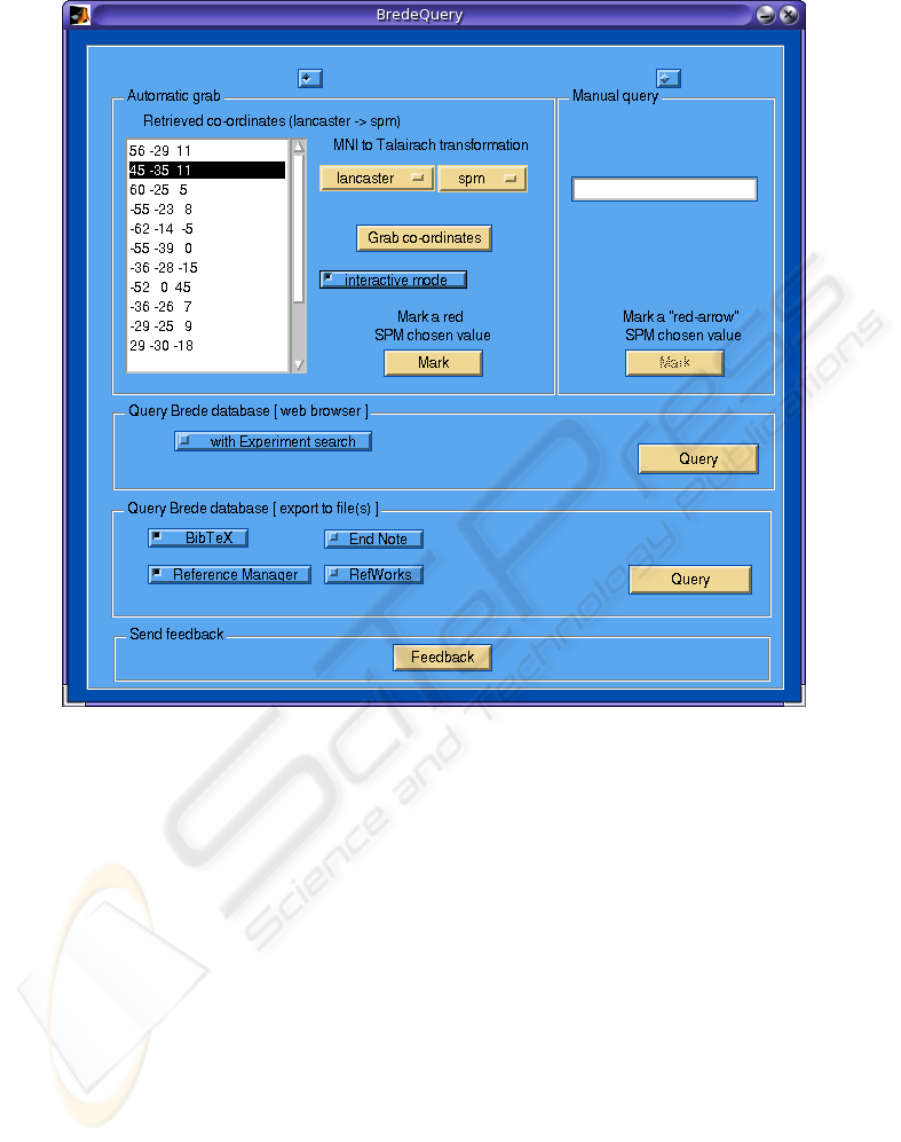
Figure 1: Graphical user interface of the BredeQuery plugin for SPM5. Firstly, the user can choose if the coordinates used
for querying will be grabbed from an SPM’s results window or will be typed manually. The grabbed (retrieved) coordinates
are shown on the list. The user can switch on an interactive mode – the coordinate selected in the SPM window will be
automatically selected in the plugin on the coordinates list. Moreover, the coordinates are grabbed using the chosen MNI to
Talairach transformation (Brett or Lancaster MTT affine transformations). Afterwards, the user is able to display the query
results in the Matlab web browser or to import them into the specified bibliographic format.
tive search for experiments, hence, scientific papers,
which report similar coordinate sets in brain. Brain-
Map (Laird et al., 2005) and Brede (Nielsen, 2003)
are the databases which offer the coordinate-based
searching. For Brede it is available on both the web-
page and in a standalone application.
In order to enable a neuroimaging scientists to per-
form meta-analysis in the context of a specific on-
going study we here propose a tool that integrates
retrieval of related research within the data analysis
pipeline. The dominant tool for human brain map-
ping is undisputable the SPM set of tools developed
and distributed by the Functional Imaging Laboratory
(London, (Friston et al., 2007)). For an analysis of the
usage of imaging pipelines see e.g., (Nielsen et al.,
2006). Thus we have initiated the development of a
plugin for SPM5, which offers high integration with
the Brede Database.
The BredeQuery plugin (see Figure 1) provides
the opportunity to perform coordinate-based query
and retrieval of the related articles references directly
from the SPM (Matlab) environment.
2 BREDE DATABASE
The Brede Database available through the web-
page: http://hendrix.imm.dtu.dk/services/jerne/brede/
records published neuroimaging experiments that list
stereotaxic coordinates in so-called MNI or Talairach
space (Talairach and Tournoux, 1988). Presently,
close to 4000 coordinates from 186 papers with a total
of 586 experiments are available.
HEALTHINF 2009 - International Conference on Health Informatics
12
Figure 2: Screenshot from one of the pages in the Brede
Database showing coordinates in Talairach space. This is
one of presently 586 experiments recorded in the database
– an fMRI experiment resulting in 29 reported coordinates.
The data is stored in XML files, and Matlab func-
tions generate static webpages with visualization of
the entries in the database, see Figure 2. Web-based
searching is possible from the homepage, but up till
now it has required that the researcher manually typed
in the query or extracted results from the image ana-
lysis program. Webpages are also available with the
Brede Database ontologies, which exist for brain re-
gions and brain functions, see Figure 3.
3 RELATED TOOLS
There a a few available tools that have aims related to
those of the BredeQuery plugin.
The AMAT SPM toolbox is also meta-analysis
toolbox (Hamilton, 2005) for Matlab based meta-
analysis of fMRI data. It provides coordinate-based
search for over 5000 coordinates from 213 published
papers of which some were derived from the Brede
Database. The coordinates are in MNI or Talairach
space. The toolbox can locate neighboring coordi-
nates to a given coordinate, as well as publications
for a given author or year. The tool was last updated
in 2005.
Another related toolbox, is xjView (Cui and
Li, 2007), which offers the SPM user, apart
from viewing the images in glass view, sec-
tion view or 3D render view, search of selected
brain regions in databases in order to eluci-
date their function. It searches among others
in Google Scholar (http://scholar.google.com/),
PubMed (http://www.ncbi.nlm.nih.gov/pubmed/)
and the xBrain database, available on the webpage:
http://sig.biostr.washington.edu/projects/xbrain/.
The XCEDE SPM Toolbox (Keator et al., 2006)
is also a toolbox distributed within the SPM com-
munity. This is a toolbox for SPM99 and SPM2,
which enables the users to capture activation data
for PET/fMRI analysis and save them to the XML
file in a XCEDE XML schema. Moreover, it is
extending the exported XML file by adding auto-
matically the anatomical labeling of the region in
the brain for the given activity coordinates. It is
achieved through two SPM toolboxes: Talairach Dae-
mon available on http://www.talairach.org/ and Au-
tomated Anatomical Labeling (AAL) available on
http://www.cyceron.fr/freeware/.
4 SOFTWARE DESCRIPTION
The recent version of the BredeQuery plugin, together
with the User’s Guide, can be downloaded from the
webpage: http://neuroinf.imm.dtu.dk/BredeQuery/. A
graphical user interface of the BredeQuery plugin is
divided into five areas where different user-actions
can be performed. Firstly, the activity coordinates
can be ‘grabbed’ from the SPM results figure into
the plugin. Since the coordinates can be presented
in MNI or Talairach spaces, some transformations are
introduced for interoperability. The coordinate-based
search in the Brede Database is based on the Talairach
space coordinates, thus the BredeQuery plugin of-
fers two MNI to Talairach transformations, which can
be chosen by the user. The piece-wise affine trans-
formation proposed by Matthew Brett is one of the
available transformation (Brett, 1999). Also included
is the affine transformation MNI-to-Talairach (MTT),
suggested by Jack Lancaster et al. (Lancaster et al.,
2007). Three separate transformation were suggested
by the group: one for SPM, one for FSL and a com-
bined ’pooled’ transformation. The MTT
SPM
trans-
formation is set as default in the BredeQuery plugin.
When the coordinates have been ‘grabbed’ and
shown in the BredeQuery plugin, the coordinate-
based querying with Brede Database can be done.
One or more coordinates can be selected for querying
and the results from the Brede Database (publications
related to the given activity coordinate) are displayed
by the plugin in a web browser (see Figure 5), ex-
ported to an XML file or saved in the bibliographic
file format (BibTeX, Reference Manager, RefWorks
or EndNote). We mention that the coordinates need
not necessarily be grabbed from SPM in order to
make a query. The coordinates can also be entered
manually in a manner similar to the functionality on
the webpage of the Brede Database.
COORDINATE-BASED META-ANALYTIC SEARCH FOR THE SPM NEUROIMAGING PIPELINE - The BredeQuery
Plugin for SPM5
13
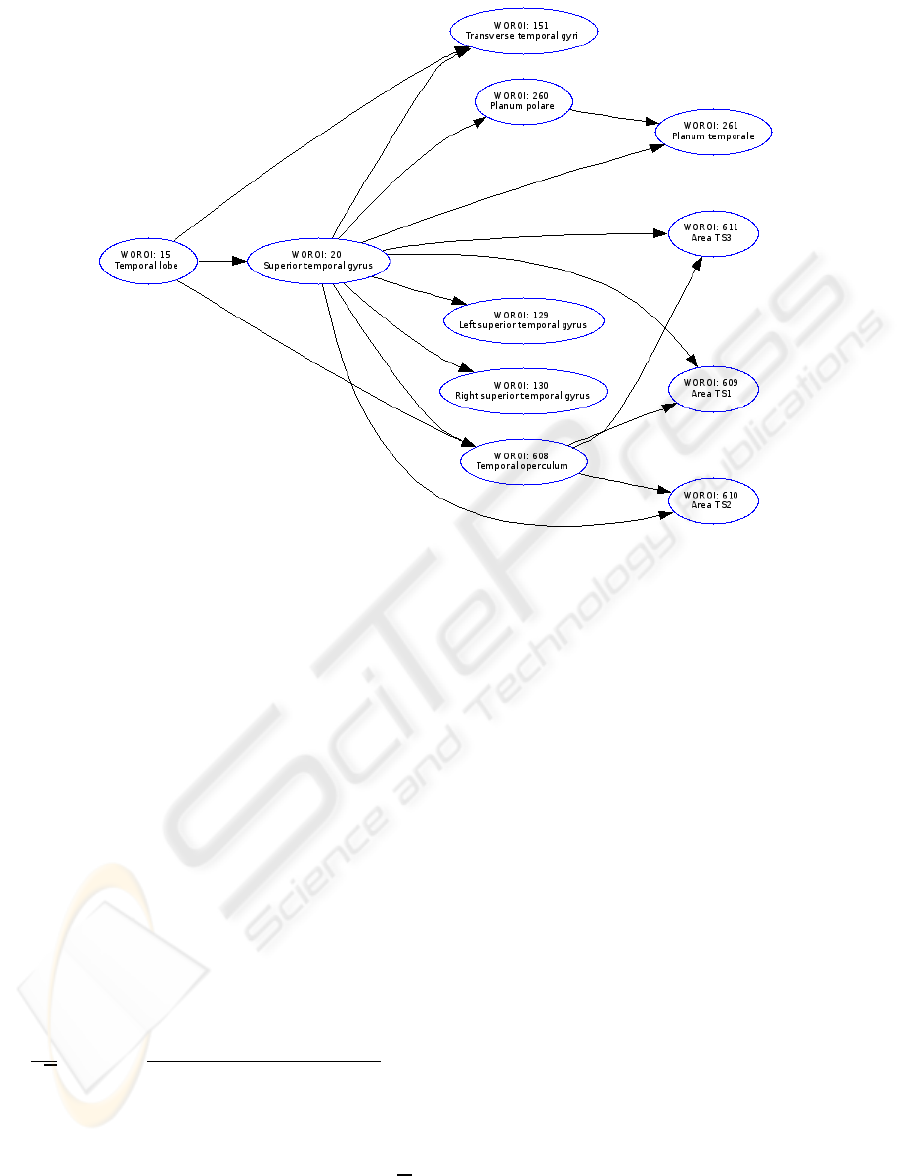
Figure 3: Relationships and taxonomy of the regions in brain associated with superior temporal gyrus. The entire ontologies
for brain regions and brain functions are available together with the Brede Database.
The user is also able to perform an ‘experi-
ment search’ (available in the Brede Database
service) via the BredeQuery. It has previously
been suggested how a similarity can be computed
between one set of coordinates and a volume or
another set of coordinates (Nielsen and Hansen,
2004). This procedure required the conversion
of the set of coordinates to a volume by kernel
density estimation. It is, however, not necessary
to convert the coordinates to a volume if only the
similarity between two coordinates sets are to be
compared. It will then generally be faster to compute
the similarity based on all coordinate-coordinate
pair-wise similarities and perform a weighted sum-
mation. There are multiple ways to compute the
similarity. Presently, the web-service for the Brede
Database uses the following Gaussian/Euclidean form
s
q,e
=
1
√
N
M
∑
m=1
N
∑
n=1
exp
(x
q
m
−x
e
n
)
2
+ (y
q
m
−y
e
n
)
2
+ (z
q
m
−z
e
n
)
2
−2σ
2
,
where σ is set to 10 millimeters, (x
q
m
,y
q
m
,z
q
m
) is the
mth of M three-dimensional query coordinates,
while (x
e
n
,y
e
n
,z
e
n
) is the nth of N three-dimensional
coordinates in the Brede Database. The factor 1/
√
N
aims to regularize for the number of coordinates in
each set so that sets with many coordinates do not
dominate the search result. A corresponding weight
for the query coordinates is not necessary, since this
factor will be equal for all queried sets of coordinates
of the database.
Following the terminology of BrainMap, a set of
coordinates is in the Brede Database called an ‘expe-
riment’ (Fox et al., 1994), thus the name ‘experiment
search’.
The Perl function that presently pro-
vides the search functionality from the Brede
Database webpage is part of the Brede Tool-
box, and this toolbox is available on the Internet
http://hendrix.imm.dtu.dk/software/brede/.
5 EXAMPLE SESSION
In this section we present the example steps for the
user session in SPM5 using BredeQuery plugin. The
sample data – Single subject epoch (block) audi-
tory fMRI activation data, taken from SPM webpage:
http://www.fil.ion.ucl.ac.uk/spm/data/auditory/, was
used for this demonstration purposes. An example
SPM5-BredeQuery session may proceed with the fol-
lowing steps:
1. In an analysis of PET/fMRI data with SPM5, the
data set is loaded, a statistical analysis is per-
HEALTHINF 2009 - International Conference on Health Informatics
14
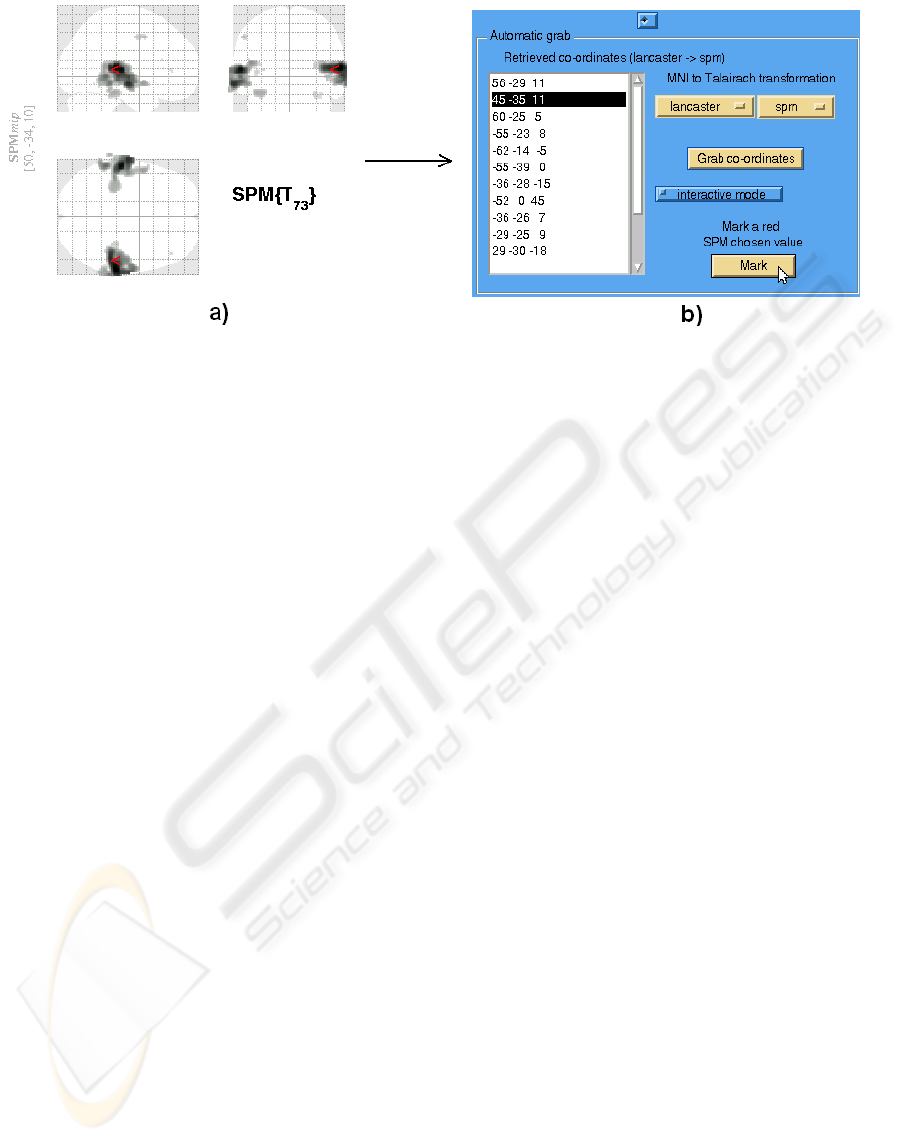
Figure 4: The demonstration of example usage of the BredeQuery plugin in SPM. The user can see regions with significant
brain activation in the SPM results window. The region of activation represented by the coordinate (50, -34, 10) in MNI space
is selected (a). The same coordinate, transformed to the Talairach space using Lancaster’s MTT
SPM
transformation is marked
on the BredeQuery’s coordinates list as (45, -35, 11) (b). Afterwards, the user is able to submit coordinate-based queries to
the Brede Database and get the articles related to the same (or nearby) brain regions.
formed and, finally, results are displayed in an
SPM window. The regions of significant brain
activation are marked and their coordinates can
be retrieved. The currently selected activation is
marked with a small red arrow. In our example
(Figure 4), the user has loaded and analyzed the
data. After the analysis, he has selected the acti-
vation, represented by the coordinate in the MNI
space as (50, -34, 10) – see Figure 4a.
2. The BredeQuery plugin is run by choosing the
BredeQuery entry in the SPM’s toolbox pop-up
menu. All coordinates are grabbed by the plugin
and shown in the coordinates list. They are trans-
formed according to the chosen MNI-to-Talairach
transformation. In our example, the coordinates
were transformed using the Lancaster’s MTT
SPM
affine transformation. The user has pressed the
Mark red SPM chosen value button and the pre-
viously selected coordinate (50, -34, 10) in MNI
space, transformed in the plugin to (45, -35, 11)
in Talairach space, is marked in the plugin’s coor-
dinates list – see Figure 4b.
3. The user has pressed the Query button in the
Query Brede database [web browser] panel
(shown on Figure 1) and the webpage with the
query results (related articles) has appeared. The
user is now able to compare the obtained results
and conclusions with those from the retrieved ar-
ticles. The webpage results from our example are
displayed on Figure 5. The first match from the
Brede Database is to a coordinate in superior tem-
poral gyrus from an experiment titled Tics during
Tourette’s syndrome. There is a link to the taxo-
nomy of the regions in brain associated with supe-
rior temporal gyrus (see Figure 3). Moreover, the
user can see the abstract to the article connected
with the above mentioned experiment.
4. The user wants to reference some of the articles
from the retrieved results in a manuscript. He
has selected the bibliographic format, which he
is going to use (in this example case ‘BibTeX’),
pressed Query button in the Query Brede database
[export to file(s)] panel (shown on Figure 1) and
the BibTeX file with the references was obtained.
5. The user has discovered a missing feature in the
BredeQuery plugin. He thus has pressed the Feed-
back button (Figure 1) and sent a comment to the
develop team.
6 FUTURE WORK
The presented plugin for SPM5 is still under develop-
ment and more features are planned.
It was recently emphasized that there are many
separated research communities in neuroscience,
which do not want to share or exchange the ex-
perimental data (Ascoli, 2006). Researchers have
expressed concerns that sharing of data can lead
to unfair use (Teeters et al., 2008). However,
data sharing is an important current issue in neu-
roscience (Kennedy, 2007; Liu and Ascoli, 2007)
and it is believed that broad data sharing could lead
to breaktroughs in our understanding of brain func-
tion (Van Horn and Ball, 2008). Invoking online
social networks and computer-based communication
can support closer relationships and trust (Lampe
COORDINATE-BASED META-ANALYTIC SEARCH FOR THE SPM NEUROIMAGING PIPELINE - The BredeQuery
Plugin for SPM5
15
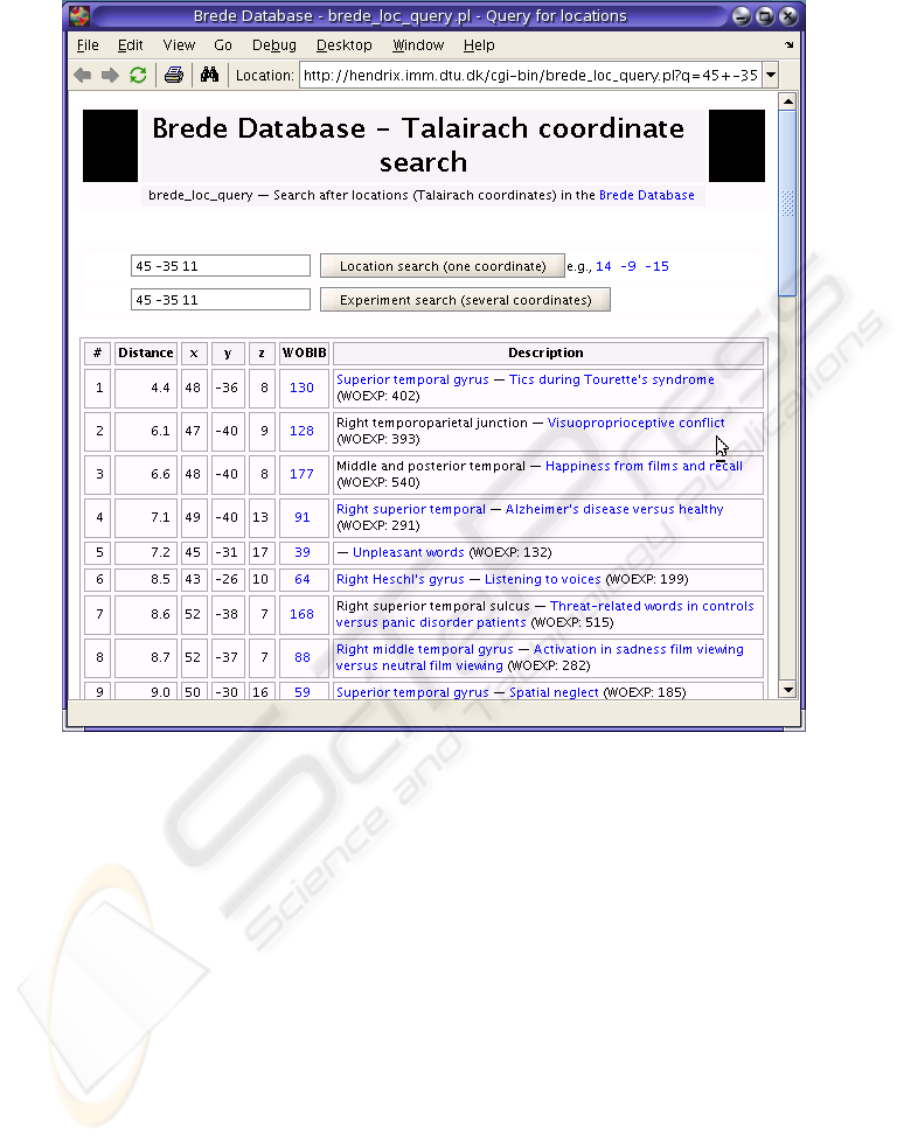
Figure 5: Brede Database query – result displayed in a web browser. List of nearby coordinates to a queried coordinate,
displaying distance, the three-dimensional coordinates, the paper identifier, the anatomical label for the retrieved coordinates
and short description of the experiment.
et al., 2006) hence, reduce the resistance to data sha-
ring.
Consequently, an interesting extension of the
functionality of the plugin can be a direct connec-
tion from the SPM environment to a neuroscientific
research community, web service or social network.
The user would be able to upload the coordinates,
results of the analysis, to his own account and save
in the assigned server disk space in order to process
them later. He can decide whether he wants to keep
it private, share only with his research group or alter-
nately release it as a public resource to all users of the
service.
It is also possible to employ the BredeQuery
plugin to expand the Brede Database. The increment
in number of the articles stored in the database could
cause bigger interest from the neuroscientists. They
could then be encouraged to register their published
or unpublished publications in the database via the
BredeQuery plugin together with the reported coor-
dinates and keywords.
7 CONCLUSIONS
In this paper we presented the BredeQuery plugin for
SPM5 - an application which offers a direct link from
SPM5 to the Brede Database. We provide a mech-
anism which allows the SPM user to find references
to articles which relate to the similar brain activation
areas through so-called coordinate-based searching.
Moreover, the BredeQuery plugin facilitates the cre-
ation of the bibliography files in popular formats.
HEALTHINF 2009 - International Conference on Health Informatics
16

ACKNOWLEDGEMENTS
We would like to thank Torben Lund and Julian Ma-
coveanu for very constructive comments and feed-
back. This work is supported by Lundbeckfonden
through the Center for Integrated Molecular Brain
Imaging (CIMBI) – www.cimbi.org.
REFERENCES
Ascoli, G. A. (September 2006). The Ups and Downs
of Neuroscience Shares. Neuroinformatics, 4:213–
216(4).
Brett, M. (1999). The MNI brain and the Talairach atlas.
MRC Cognition and Brain Sciences Unit.
Cui, X. and Li, J. (2007). xjView - a viewing program for
SPM. http://people.hnl.bcm.tmc.edu/cuixu/xjView/.
Fox, P. T., Mikiten, S., Davis, G., and Lancaster, J. L.
(1994). BrainMap: A database of human function
brain mapping. In Thatcher, R. W., Hallett, M., Zef-
firo, T., John, E. R., and Huerta, M., editors, Func-
tional Neuroimaging: Technical Foundations, chap-
ter 9, pages 95–105. Academic Press, San Diego, Cal-
ifornia.
Friston, K., Ashburner, J., Kiebel, S., Nichols, T., and
Penny, W., editors (2007). Statistical Parametric
Mapping: The Analysis of Functional Brain Images.
Academic Press.
Hamilton, A. (2005). AMAT - a meta-analysis toolbox.
http://www.antoniahamilton.com/amat.html.
Keator, D. B., Gadde, S., Grethe, J. S., Taylor, D. V., and
Potkin, S. G. a. (2006). A general XML schema and
SPM toolbox for storage of neuro-imaging results and
anatomical labels. Neuroinformatics, 4(2):199–212.
Kennedy, D. N. (2007). Neuroinformatics and the Society
for Neuroscience. Neuroinformatics, 5:141–142.
Laird, A. R., Lancaster, J. L., and Fox, P. T. (2005). Brain-
Map: The Social Evolution of a Human Brain Map-
ping Database. Neuroinformatics, 5:65–78.
Lampe, C., Ellison, N., and Steinfield, C. (2006). A
face(book) in the crowd: social searching vs. social
browsing. In CSCW ’06: Proceedings of the 2006
20th anniversary conference on Computer supported
cooperative work, pages 167–170, New York, NY,
USA. ACM Press.
Lancaster, J. L., Tordesillas-Guti´errez, D., Martinez, M.,
Salinas, F., Evans, A., Zilles, K., Mazziotta, J. C., and
Fox, P. T. (2007). Bias between MNI and Talairach
coordinates analyzed using the ICBM-152 brain tem-
plate. Human Brain Mapping.
Liu, Y. and Ascoli, G. A. (September 2007). Value Added
by Data Sharing: Long-Term Potentiation of Neuro-
science Research: A Commentary on the 2007 SfN
Satellite Symposium on Data Sharing. Neuroinfor-
matics, 5:143–145(3).
Nielsen, F.
˚
A. (2003). The Brede database: a small database
for functional neuroimaging. In NeuroImage, vol-
ume 19. Elsevier. Presented at the 9th International
Conference on Functional Mapping of the Human
Brain, June 19-22, 2003, New York, NY.
Nielsen, F.
˚
A., Christensen, M. S., Madsen, K. M., Lund,
T. E., and Hansen, L. K. (2006). fMRI Neuroinfor-
matics. IEEE Engineering in Medicine and Biology
Magazine, 25(2):112–119.
Nielsen, F.
˚
A. and Hansen, L. K. (2002). Modeling of acti-
vation data in the BrainMap(TM) database: Detection
of outliers. Human Brain Mapping, 15(3):146–156.
Nielsen, F.
˚
A. and Hansen, L. K. (2004). Finding related
functional neuroimaging volumes. Artificial Intelli-
gence in Medicine, 30(2):141–151.
Nielsen, F.
˚
A., Hansen, L. K., and Balslev, D. (2004). Min-
ing for associations between text and brain activation
in a functional neuroimaging database. Neuroinfor-
matics, 2(4):369–380.
Pekar, J. (2006). A brief introduction to functional MRI.
IEEE Engineering in Medicine and Biology Maga-
zine, 25(2):24–26.
Talairach, J. and Tournoux, P. (1988). Co-planar Stereo-
taxic Atlas of the Human Brain. Thieme Medical Pub-
lisher Inc, New York.
Teeters, J. L., Harris, K. D., Millman, K. J., Olshausen,
B. A., and Sommer, F. T. (2008). Data Sharing for
Computational Neuroscience. Neuroinformatics.
Van Horn, J. D. and Ball, C. A. (2008). Domain-Specific
Data Sharing in Neuroscience: What Do We Have to
Learn from Each Other? Neuroinformatics.
Wager, T. D., Lindquist, M., and Kaplan, L. (2007). Meta-
analysis of functional neuroimaging data: current and
future directions. Social Cognitive and Affective Neu-
roscience, 2(2):150–158.
COORDINATE-BASED META-ANALYTIC SEARCH FOR THE SPM NEUROIMAGING PIPELINE - The BredeQuery
Plugin for SPM5
17
