
AN EXTENSIBLE BIOMARKER CURATION APPROACH AND
SOFTWARE INFRASTRUCTURE FOR THE EARLY DETECTION
OF CANCER
Andrew F. Hart
1
, John J. Tran
1,3
, Daniel J. Crichton
1
, Kristen Anton
2
Heather Kincaid
1
, Sean Kelly
1
, J. S. Hughes
1
and Chris A. Mattmann
1,3
1
Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA 91109, U.S.A.
2
Section of Biostatistics and Epidemiology, Dartmouth Medical School, Lebanon, NH 03766, U.S.A.
3
Computer Science Department, University of Southern California, Los Angeles, CA 90089, U.S.A.
Keywords:
Bioinformatics, Data grid, Data management, Data procurement, Ontology.
Abstract:
Modern research requires collaboration among geographically distributed scientists. This collaborative model
is transforming scientific discovery by enabling sharing and validation of data across institutions. Informatics
infrastructures are being developed to support cancer research, endowing scientists with the ability to capture
and share data with remote collegues. A critical challenge presented by such infrastructures is the develop-
ment of a curation model for the science data. While considerable emphasis has been placed on developing
grid infrastructures, few are addressing the curation aspects crucial to creating a useful scientific knowledge-
base. The United States National Cancer Institute’s (NCI) Early Detection Research Network (EDRN) is a
distributed network of research institutions focused on the discovery of cancer biomarkers. In this paper, we
describe our work building a data collection and curation infrastructure on top of the existing EDRN bioinfor-
matics data grid. The approach involves normalizing curated data through the use of a common information
model for cancer biomarker research. We argue that such a model is critical to ensuring that data can be com-
bined into an integrated knowledge system. Furthermore, we argue that human curators with backgrounds in
both informatics and science play a critical role in the overall value of the EDRN knowledge-base.
1 INTRODUCTION AND
MOTIVATION
In recent years, the cancer research community has
seen increasing dependency on complex computing
infrastructure (clusters, exotic instrument technolo-
gies with associated software and data formats, etc.)
and data modeling ‘know-how’ (common data ele-
ments, ontologies, etc.) to support its scientific re-
search needs. In contrast to the silo’ed research labs
of old, scientists are now under immense pressure
to exchange, share and disseminate data across geo-
graphically dispersed institutions. As has been shown
(Crichton et al., 2006), the rate of scientific discovery
increases as data is shared between collegues. The
need to share data has necessitated a common vocab-
ulary not only for the format of data (the organization
of its “bits”), but for annotations about the data itself
– regularly referred to as metadata.
We have borne witness first-hand to the prolifera-
tion of data management needs specific to the cancer
research community, as have others more generally
within the field of biology (Lynch, 2008; Howe et al.,
2008). Over the past seven years, we have helped to
construct and deploy the enabling grid infrastructure
(Foster et al., 2001) for a highly successful bioinfor-
matics grid project within the U.S. National Cancer
Institute (NCI). The Early Detection Research Net-
work (EDRN) project is a network of over forty insti-
tutions across the U.S. all collaborating with the com-
mon goal of the early detection of cancer through dis-
covery of promising cancer biomarkers – predictors
of cancer at an early stage.
To assist EDRN in the curation of its many types
387
F. Hart A., J. Tran J., J. Crichton D., Anton K., Kincaid H., Kelly S., S. Hughes J. and A. Mattmann C. (2009).
AN EXTENSIBLE BIOMARKER CURATION APPROACH AND SOFTWARE INFRASTRUCTURE FOR THE EARLY DETECTION OF CANCER.
In Proceedings of the International Conference on Health Informatics, pages 387-392
DOI: 10.5220/0001781103870392
Copyright
c
SciTePress

of data, we have built a suite of applications col-
lectively termed the EDRN Knowledge Environment
(EKE) (Crichton et al., 2006). EKE consists of:
a distributed specimen inventory called ERNE (for
EDRN Resource Network Exchange), study manage-
ment database applications called VSIMS and eSIS,
and two technologies that we will focus on in this pa-
per: the Biomarker Database and the EDRN Catalog
and Archive Service (or eCAS for short). Each appli-
cation is an independent software component respon-
sible for capturing and annotating its particular type
of EDRN data and/or metadata. Periodically (or at
defined intervals), metadata from each of these ap-
plications can be pushed a centralized EDRN web
portal that accepts Resource Description Framework
(RDF) (Lassila and Swick, 1999) exports of the data
and metadata captured in each application, for use in
specialized search and discovery user interfaces such
as free-text and facet-based search.
Our recent work involves developing a stan-
dard procedure for curating information in both the
Biomarker Database and the eCAS. Curation of high
quality, peer-reviewed information for each of these
applications has numerous benefits within the EDRN,
including: (1) providing a means for tracking research
publications associated with discovered biomarkers
(an indicator of viability of promising biomarkers
identified by EDRN investigators); (2) for track-
ing biomarker validation (EDRN has a five phase
biomarker validation process ranging from early in-
vestigation to clinically validated results); and (3) for
sharing and disseminating both raw and processed
science data of use to researchers in the biomedical
community.
In this paper, we discuss our experience in devel-
oping a data model and curation approach and asso-
ciated software infrastructure for curating biomarker
information from the Biomarker Database and eCAS.
Though in its nascent stages, the curation approach
and software are proving worthwhile as we have been
successful in capturing publication, study, organ and
sensitivity/specificity information for a number of
promising biomarkers in the EDRN, and also for a
number of popular science data sets.
The rest of this paper is organized as follows. Sec-
tion 2 discusses background and related work in the
areas of bioinformatics grid infrastructures and data
curation for biomedicine. Section 3 presents our data
model, approach, and software infrastructure for cu-
rating biomarker research information, and important
cancer science data sets. Section 4 describes the use
of our curation system and approach in the context of
real EDRN use cases. Section 5 rounds out paper by
pointing the reader to future work and conclusions.
2 RELATED WORK
In this section we discuss relevant related work in the
areas of large-scale bioinformatics grid infrastructure
and data curation for cancer research, highlighting
similar projects and underpinning the unique contri-
bution of our own work – the assertion that, unlike
related projects, data curation has been made a first-
class process within our work within the context of
curating biomarker information in the EDRN.
2.1 Bioinformatics Grid Infrastructure
There are several related projects building bioinfor-
matics grid infrastructure. We discuss a representative
subset of them here.
2.1.1 BIRN
The Biomedical Informatics Research Network
(BIRN) is a comparable initiative being undertaken by
the National Center for Research Resources (NCRR)
and the National Institutes of Health (NIH) for the
purpose of sharing neuroscience and brain imaging
data between collaborating scientists (Keator et al.,
2008). Like the EDRN, BIRN participants conduct
research at their own facilities and retain control over
the data they generate. While clearly a successful
project, BIRN allows scientists to submit data via the
BIRN portal, according to a specification documented
in an associated PDF file (BIR, 2008). Our experience
indicates that allowing scientists to curate their own
data, unmoderated, can at times introduce data qual-
ity and overlap issues that can be difficult to resolve.
Within EDRN, we are focused on curation by highly
trained informatics personnel, with science expertise
in a particular organ-related cancer.
2.1.2 caBIG
The Cancer Biomedical Informatics Grid (caBIG)
(von Eschenbach and Buetow, 2006) is a large, ini-
tiative funded by the U.S. National Cancer Institute
(NCI) whose mission includes building out an all-
inclusive bioinformatics grid infrastructure for shar-
ing data among the whole of the cancer research com-
munity. Like BIRN, caBIG embraces the idea that its
participating sites maintain control of the data they
contribute.
A primary difference between the EDRN and
caBIG initiatives is their respective scope. Rather
than endeavoring towards a broad-reaching connec-
tive web for cancer research as a whole, the EDRN
has taken a more refined approach, focusing narrowly
in on the specific issues facing researchers involved
HEALTHINF 2009 - International Conference on Health Informatics
388

in the early detection of cancer. We have recently in-
tegrated EDRN with a caBIG enabled specimen cu-
ration tool, caTissue, which we will discuss in detail
below.
2.2 Curation of Biomarker Information
While there are several tools and efforts already un-
derway to capture biomarker related medical data
(e.g., see (Keerthi et al., 2002; Baral et al., 2005)),
due to space limitations we will focus on two re-
lated approaches for curating specimen information
called caTissue and the National Biospecimen Net-
work (NBN), respectively.
2.2.1 caTissue
caTissue (caT, 2008), is a specimen capture system
developed at the NCI. Focusing largely on gene ex-
pression and sequence data as it relates to cancer re-
search, the system operates as a ’plug-in’ to caBIG.
Patient tissue sample is combined with metadata an-
notations to form a comprehensive specimen bank
that can be queried by a caBIG user.
It should be noted that ERNE has successfully
plugged into the caTissue suite, and can thus EDRN
can integrate with caBIG grids.
2.3 NBN
The National Biospecimen Network (NBN) (Birm-
ingham, 2004) is an NCI-funded initiative arising out
of a general consensus during the March 2002 Na-
tional Dialogue on Cancer that access to annotated tis-
sue collections was a critical enabling factor in the ap-
plication of recent technological advances to the fight
against cancer. NBN has faced a similar need for a
framework in which to provide meaningful annota-
tions of the tissue samples. While the scope of the
NBN is much more broadly defined than the EDRN,
there is some degree of overlap with regards to data
curation issues.
3 CURATION OF BIOMARKER
DATA AND RAW SCIENCE
RESULTS
While it is undeniable that a robust, extensible infras-
tructure is an integral part of a complete informatics
solution, we feel that the issues surrounding the cu-
ration of that data need to be placed on at least equal
footing. The specialized nature of the data generated
by cancer biomarker research, combined with issues
of coordination and control that arise from the inher-
ent geographical distribution of the work being done,
place an emphasis on the importance of data curation.
In this section we will discuss our approach to build-
ing data models and curation tools for the BMDB and
eCAS.
3.1 Biomarker Database Data Model
The raw input data for the Biomarker Database is pro-
vided by various distributed applications and services.
Interoperability with these data sources depends on
the collective ability of all participants to ‘speak the
same language.’ To that end, the Biomarker Database
relies heavily on a common information model, the
EDRN Ontology, used throughout EDRN informat-
ics. The Ontology provides all applications with
common reference for naming conventions using the
EDRN registry of Common Data Elements (CDE), as
well as relationships that add meaning between the
various biomarker research data objects. The EDRN
Ontology has been an organizing presence at every
step in the development of the BMDB, from the de-
sign of the initial ingestion and storage routines, to
specifications for the export of the curated data. Most
critically, however, the Ontology provides a formal
foundation for the design of the BMDB data model.
The EDRN Ontology has been implemented using
Stanford’s Prot
´
eg
´
e toolkit (Noy et al., 2000)
Because the data is so highly specialized, the data
model for the BMDB needs to be expressive and flex-
ible, providing a curator with the ability to indicate
associations between data objects with as much lati-
tude as possible. Research on a particular biomarker
is comprised of many individual elements, many of
which may be related on sometimes overlapping lev-
els. As an example, a given study may make a passing
reference to a biomarker as a member of a panel of
biomarkers. Alternatively, a study might consider the
biomarker’s relationship to a particular organ-site in
great detail. Each of these studies may have relevant
publications, external resources, and other important
relationships to the biomarker that need to be curated
in order to truly capture a comprehensive representa-
tion of the research.
The challenge has been to develop a data model
flexible enough to handle these nuances, yet refined
and focused enough to retain the semantic meaning
of the underlying information. Figure 1a indicates
the quantity of supporting data that can be associated
with a biomarker by a curator. It is important to note
that a given type of data (a publication, for example)
can be associated at a number of locations, reflecting
AN EXTENSIBLE BIOMARKER CURATION APPROACH AND SOFTWARE INFRASTRUCTURE FOR THE
EARLY DETECTION OF CANCER
389
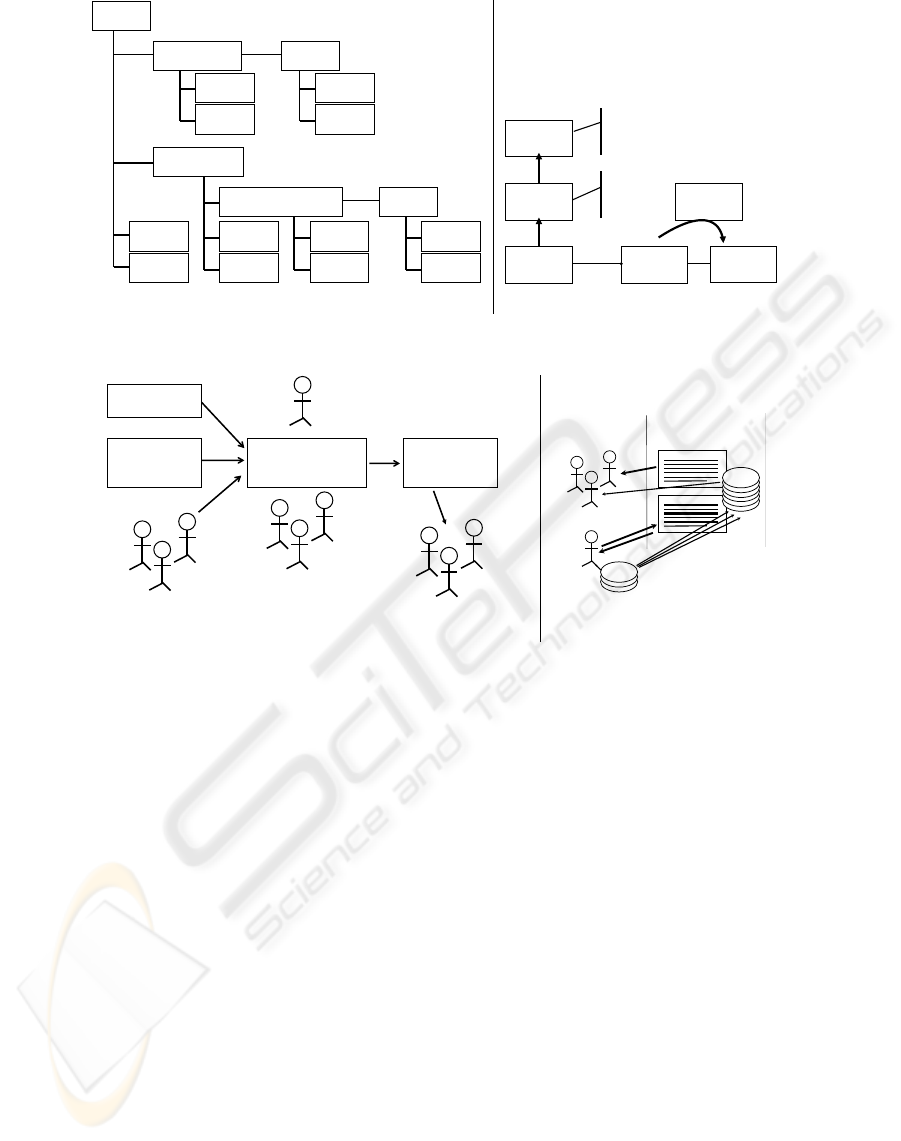
Biomarker
Biomarker-Study
Data
Study
Publication
Resource
Biomarker-Organ
Data
Biomarker-Organ-Study
Data
Study
Publication
Resource
Publication
Resource
Publication
Resource
Publication
Resource
Publication
Resource
CAS File
EDRN
Data Set
SELDI
product id
filename
product type
file location
...
study
specimen
...
productType elements
type-element
map
instance of
(a) (b)
Figure 1: (a) Biomarker Database data model diagram and (b) example eCAS data model.
Biomarker
Database
EDRN Public
Portal
DMCC Protocol
Database
PubMed
Publication
Repository
EDRN P.I.s
Peer-Review Board
Curator
Public
RDF
XML
HTTP
RDF
HTTP
users: read only
curator: read & write
eCAS system
high fidelity data
ingestion
meta data
(a) (b)
Figure 2: (a) Biomarker Database process flow and (b) eCAS data curation flow.
the flexibility a curator has in indicating data relation-
ships. Likewise, it is also possible to link a particular
study with a biomarker in meaningful ways: (1) re-
lated directly to the biomarker without mentioning a
specific organ (or, alternatively, mentioning multiple
organs); or (2) as specifically relating the study to a
given biomarker-organ pair. We have found that pro-
viding such a meaningful variety of curation options
greatly enhances the utility of the curated data.
3.2 Biomarker Database Curation
Process
The Biomarker Database curation process is realized
through a combination of efforts of one or more cura-
tors with a background in both informatics and biol-
ogy, and supporting software tools (as shown in Fig-
ure 2a). Active communication between researchers
and a dedicated curator serves to mitigate the difficul-
ties posed by the ad-hoc submission of research data
from multiple independent sites. The curator acts as
a liaison between the research scientists and the sys-
tem, providing an additional layer of assurance on the
quality of the data.
Once a biomarker has been selected for curation,
the curator works with the research team to identify
the relevant details (including EDRN studies, sensi-
tivity, specificity, and predictive value data, publica-
tions and resources) that would contribute to a greater
understanding of the current research state. Using a
streamlined, web-based tool, a curator is able to indi-
cate associations between the data, directly importing
and linking resources from across the EDRN enter-
prise as needed to create a unified view of the research
that is ready for the peer review process.
3.3 eCAS Data Model
Whereas the Biomarker Database data model has
been designed to specifically store research data
related to the discovery and validation of cancer
biomarkers, the eCAS data model is generalizable to
all scientific research communities. The model it-
self places no restrictions on the type of data being
processed, and follows an inheritance pattern which
allows for unlimited extensibility and specialization.
The core meta-data representation consists of three
HEALTHINF 2009 - International Conference on Health Informatics
390

basic building blocks: (1) elements – basic key/pair
nucleus; (2) productType – a composite set of ele-
ments; and (3) typeElementMap – mapping represen-
tation between a product type definition and its corre-
sponding elements list.
All meta-data representations are defined using
these three core data structures. More complex data
definitions specific to the particular science domain
inherit and extend their properties from the base-
line definition. This built-in extensibility allows an
eCAS instance to be tailored to suit even very com-
plex data warehousing scenarios. Figure 1b shows
how this hiearchical organization of the eCAS data
model is extended to a specific implementation of
e.g., a Surface-enhanced laser desorption/ionization
(SELDI) dataset definition. It is possible to trace the
inheritance graph by starting with the casFile pol-
icy entity, a generic meta-data definition, and travers-
ing backwards, each entity expanding and specializ-
ing the previous one, until arriving at a policy specific
to the SELDI data set.
3.4 eCAS Curation Process
As mentioned earlier, the eCAS data model is de-
signed to be domain agnostic. As a consequence, the
web-based curation interface, modeled in Figure 2b,
does not speak to any specific data flow and its users’
roles and delegation of authority are more generic.
While the curation requirements of a particular do-
main may vary to some extent, all curation efforts in-
volve, at a minimum, (1) high-fidelity data ingestion,
and (2) meta-data manipulation.
Addressing the first concern, eCAS provides an
interface to support data ingestion. Because the eCAS
curation application is built on top of the underlying
OODT CAS layer (Mattmann et al., 2008), it is able
to support voluminous and high fidelity data inges-
tion. For the second concern, meta-data manipulation,
eCAS provides a web-based interface (similar to the
BMDB curation web interface described earlier) that
presents curators with an efficient method for entering
and editing meta-data. The tool provides an option to
update existing data (and meta-data) or initiate new
product type entry from scratch using a “wizard” in-
terface.
4 APPLICATION WITHIN EDRN
4.1 An Emphasis on Data Quality
The efforts of the EDRN center around the collec-
tion, storage, annotation and presentation of cancer
biomarker research. The BMDB and eCAS facili-
tate these efforts by providing a complete data storage
and curation infrastructure. As EDRN aims to pro-
vide authoritative, comprehensive coverage of can-
cer biomarker discovery and validation research, the
quality of the data is a paramount concern.
As discussed in Section 3, the majority of the de-
velopment effort on the Biomarker Database has been
directed towards working with the NCI, EDRN Prin-
cipal Investigators, and curators to develop a flexible
web-based curation interface to support curation ef-
forts. The collaborative effort has at times unearthed
significant model differences among the participants
which, if left undiscovered, might lead to conflicting
interpretations and ambiguities and threaten to under-
mine the value of the data.
The Biomarker Database has provided us with a
sandbox of sorts in which to iteratively refine our data
model until a consensus was reached. At present, we
have approximately 15 highly curated biomarkers and
11 data sets (including 1000s of data files and meta-
data files) representing a range of sub-disiplines in-
cluding liver, lung and ovary. Each of the biomark-
ers is meaningfully linked to studies, organ-site data,
publications, and external resources and thus forms a
largely complete picture of the existing research.
Curation is an essential ingredient in our efforts to
construct the foundations of a comprehensive knowl-
ege environment of value to the scientific community.
The presence of a knowledgeable curator capable of
acting as liaison and proverbial “traffic-cop”, has been
pivotal in maintaining the integrity of the captured
data.
4.2 Discussion
Curation is the linchpin in a process that extends from
initial ingestion of data from various external sources,
including the EDRN Data Management and Coor-
dinating Center (DMCC) Protocol Database at the
Fred Hutchinson Cancer Research Center (FHCRC)
in Seattle, and publication repositories like PubMed
(both shown in the upper left portion of Fig. 1a) to
the eventual release of peer-reviewed biomarker re-
search through the EDRN Public Portal (shown in the
middle right portion of Fig. 1a). A similar process
is shown for the eCAS as depicted in Fig. 1b. The
Biomarker Database and the eCAS system can per-
haps be thought of as data refineries, taking in vol-
umes of raw, uncorrelated data. The resulting curated,
peer-reviewed data that is ultimately made available
for public consumption (shown in the lower right por-
tion of Fig. 1a and upper left version of Fig. 1b) is of
high quality (as it has been peer-reviewed) and highly
AN EXTENSIBLE BIOMARKER CURATION APPROACH AND SOFTWARE INFRASTRUCTURE FOR THE
EARLY DETECTION OF CANCER
391

valuable (as it has been curated) as an authoritative
information resource.
The key factor distinguishing our work from that
of others building bioinformatics grids is that many
of the other bioinformatics grid efforts are pursuing
technology research and have, we believe, not given
curation sufficient prominence, even as the data man-
agement problems in science have continued to grow.
Our key lesson learned is that scientists need to be in-
volved in the planning and curation process and the
bioinformatics grid software needs to be able to grow
and evolve in unison with the data model during cu-
ration activities.
5 CONCLUSIONS AND FUTURE
WORK
In this paper we discussed our efforts to define a cura-
tion process for biomarker information collected with
the National Cancer Institute (NCI)’s Early Detection
Research Network (EDRN) project. Biomarker re-
search data is curated and stored within two appli-
cations running on top of EDRN’s data grid infras-
tructure: (1) the Biomarker Database (BMDB), and
(2) the EDRN Catalog and Archive Service (eCAS).
We described the data model and curation process for
each of these applications and described real EDRN
use cases for each application. We have experi-
enced firsthand some of the difficulties in transform-
ing raw research data from geographically diverse
sources into a comprehensive query-driven knowlege-
base. These difficulties reinforce the notion that (1)
data models must be developed with evolvability as
a cornerstone; (2) scientists need to be actively in-
volved in the model development process to the great-
est extent possible; and (3) data management (cura-
tion) policy development is at least as important to
address as decisions about the underlying technology
infrastructure.
ACKNOWLEDGEMENTS
This effort was supported by the Jet Propulsion Lab-
oratory, managed by the California Institute of Tech-
nology under a contract with the National Aeronautics
and Space Administration. The authors would like to
thank Donald Johnsey, Christos Patriotis, and Sudhir
Srivastava and the NCI leadership as a whole for their
collaborative guidance and support.
REFERENCES
(2008). Birn - describing your data,
http://nbirn.net/bdr/study information.shtm.
(2008). catissue core,
https://cabig.nci.nih.gov/tools/catissuecore.
Baral, C., Davulcu, H., Nakamura, M., Singh, P., Tari, L.,
and Yu, L. (2005). Collaborative curation of data from
bio-medical texts and abstracts and its integration. In
Data Integration in the Life Sciences, pages 309–312.
Birmingham, K. (2004). An inauspicious start for the
us national biospecimen network. J. Clin. Invest.,
113(3):320–320.
Crichton, D., Kelly, S., Mattmann, C., Xiao, Q., Hughes,
J. S., Oh, J., Thornquist, M., Johnsey, D., Srivastava,
S., Essermann, L., and Bigbee, W. (2006). A dis-
tributed information services architecture to support
biomarker discovery in early detection of cancer. In
e-Science, page 44.
Foster, I., Kesselman, C., and Tuecke, S. (2001). The
anatomy of the grid: Enabling scalable virtual organi-
zations. J. Supercomputing Applications., pages 1–25.
Howe, D., Costanzo, M., Fey, P., Gojobori, T., Hannick, L.,
Hide, W., Hill, D. P., Kania, R., Schaeffer, M., Pieer,
S. S., Twigger, S., White, O., and Rhee, S. Y. (2008).
Big data: The future of biocuration. Nature, 455:47–
50.
Keator, D., Grethe, J., Marcus, D., Ozyurt, B., Gadde,
S., Murphy, S., Pieper, S., Greve, D., Notestine, R.,
Bockholt, H., and Papadopoulos, P. (2008). A na-
tional human neuroimaging collaboratory enabled by
the biomedical informatics research network (birn).
IEEE Trans. Information Technology in Biomedicine,
12(2):162–172.
Keerthi, S. S., Ong, C. J., Siah, K. B., Lim, D. B. L., Chu,
W., Shi, M., Edwin, D. S., Menon, R., Shen, L., Lim,
J. Y. K., and Loh, H. T. (2002). A machine learn-
ing approach for the curation of biomedical litera-
ture: Kdd cup 2002 (task 1). SIGKDD Explor. Newsl.,
4(2):93–94.
Lassila, O. and Swick, R. (1999). Resource description
framework (rdf) model and syntax specification. Tech-
nical report, W3C.
Lynch, C. (2008). Big data: How do your data grow? Na-
ture, 455:28–29.
Mattmann, C., Freeborn, D., Crichton, D., Hughes, J. S.,
Ramirez, P., Hardman, S., Woollard, D., and Kelly, S.
(2008). Transformation of oodt cas to perform larger
tasks. NASA Tech Briefs., 32(6):44.
Noy, N. F., Fergerson, R. W., and Musen, M. A. (2000).
The knowledge model of protege-2000: Combining
interoperability and flexibility. In Knowledge Engi-
neering and Knowledge Management Methods, Mod-
els and Tools, pages 69–82.
von Eschenbach, A. C. and Buetow, K. (2006). Cancer in-
formatics vision: cabig. Cancer Informatics, 2:22–24.
HEALTHINF 2009 - International Conference on Health Informatics
392
