
AN ONTOLOGY FOR SUPPORTING CLINICAL RESEARCH
ON CERVICAL CANCER
Manolis Falelakis
1,2
, Christos Maramis
1,2
, Irini Lekka
3
, Pericles A. Mitkas
1,2
and Anastasios Delopoulos
1,2
1
Department of Electrical and Computer Engineering, Aristotle University of Thessaloniki, Greece
2
Informatics and Telematics Institute, Centre for Research and Technology Hellas, Greece
3
School of Medicine, Aristotle University of Thessaloniki, Greece
Keywords:
Medical ontology, Cervical cancer, Domain modelling.
Abstract:
This work presents an ontology for cervical cancer that is positioned in the center of a research system for
conducting association studies. The ontology aims at providing a unified ”language” for various heterogeneous
medical repositories. To this end, it contains both generic patient-management and domain-specific concepts,
as well as proper unification rules. The inference scheme adopted is coupled with a procedural programming
layer in order to comply with the design requirements.
1 INTRODUCTION
Medical Knowledge, being the compilation of many
years of research, has grown vast both in volume
and in complexity. Recently, the need for employing
semantically-aware models of medical knowledge
has become evident. Since then, ontologies have
been successfully used in various medical domains,
disciplines and even aspects of medical practice
and research. Examples of this include the Gene
Ontology (Ashburner et al., 2000), the ReMINE
ontology for adverse events (http://www.remine-
project.eu/), the Ontology of Clinical Re-
search (http://rctbank.ucsf.edu/home/ocre.html),
the Ontology for Clinical Investigators
(http://www.bioontology.org)etc.
This paper presents an ontology designed to
facilitate research in the domain of cervical cancer
(CxCa). This is, to the best of our knowledge, the
first ontology to deal with concepts of the CxCa
domain. It’s main targets are (i) to provide a means
for unification of various result coding formats and
(ii) to extract implicit knowledge in order to produce
potential query terms as inferred types of individuals.
Through this process, medical researchers are given
the ability to form patient groups, large enough to
provide statistically significant results in association
studies.
The paper is structured as follows: In section 2
we describe the problem, while in section 3 we try
to provide some intuition about the CxCa domain.
Section 4 is devoted to the description of the basic
structure of the proposed ontology, section 5 outlines
the medical unification rules and section 6 the infer-
encing procedure followed. Section 7 contains the
technologies utilized for implementation and some
ontology statistics and section 8 concludes the paper.
2 PROBLEM STATEMENT
In an effort to gain a more comprehensive and holis-
tic insight on the origin of complex diseases, genetic
association studies (Hirschhorn and Daly, 2005) con-
stitute a significant approach for clinical researchers.
In order to perform statistically meaningful associ-
ation studies, large amounts of clinical data are re-
quired - especially when performing studies among
many study factors. However, the current clinical
practice of constructing disposable and isolated clin-
ical research repositories hinders the construction of
collections big enough to facilitate the execution of
complex association studies. To tackle this problem,
comes the unification of existing medical repositories
that contain heterogeneous cervical cancer related in-
formation.
The need to resolve this heterogeneity, makes the
ontological representation of the CxCa knowledgethe
perfect candidate solution to this type of problems.
103
Falelakis M., Maramis C., Lekka I., A. Mitkas P. and Delopoulos A. (2009).
AN ONTOLOGY FOR SUPPORTING CLINICAL RESEARCH ON CERVICAL CANCER.
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development, pages 103-108
DOI: 10.5220/0002265701030108
Copyright
c
SciTePress
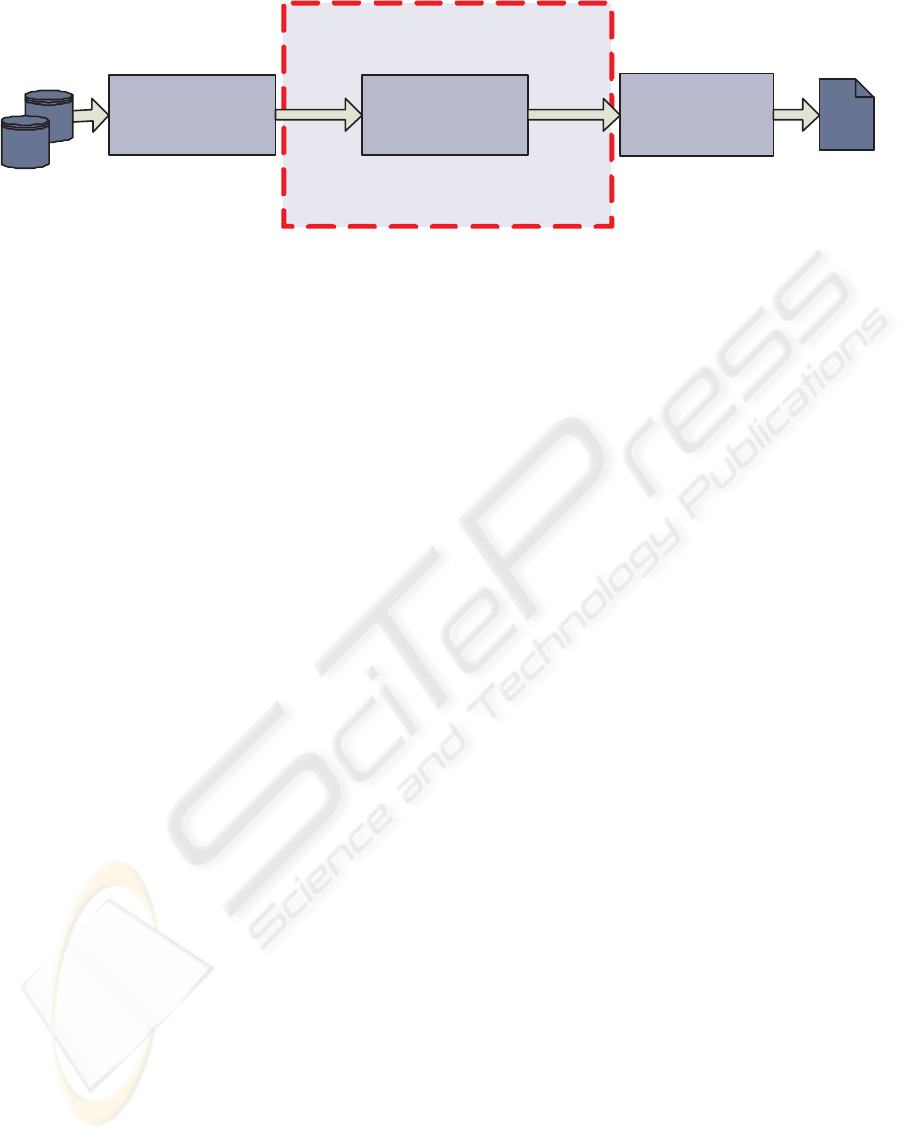
CxCa Ontology
Data Mapping
(Syntactic Unification)
Inference
(Semantic Unification)
Association Study &
Statistical Analysis
Legacy
Archives
Asserted
Knowledge
Inferred
Knowledge
Results
Figure 1: The Context of use of CxCa Ontology.
Ontologies have been widely used as unifying
models to deal with heterogeneity issues of legacy
archives. Furthermore, reasoning on top of ontologies
is a very active field of ongoing research that has al-
ready produced fruitful and concrete results (Baader
et al., 2006), (Tsarkov and Horrocks, 2006), (Hus-
tadt et al., 2004), (Dyckhoff, 2000), (Sirin et al.,
2007). Finally, there are plenty of semantic querying
languages suitable for querying on ontologies (Jeen
et al., 2004).
The ontology presented here has been developed
within the context of ASSIST (Mitkas et al., 2008),
a research project funded by the European Commis-
sion. The main objective of this project is the seman-
tic unification of physically isolated and heteroge-
neous medical repositories that include cervical can-
cer related data into one semantic repository in order
to facilitate the execution of association studies. In
this context, the CxCa ontology was built to serve as
the schema of a ’container’ repository that enables the
above unification.
The resulting data are employed by the project to
perform association studies in an automatic way. The
context of use of the CxCa Ontology is depicted in
Figure 1.
3 DOMAIN BACKGROUND
In this section we present some basic notions regard-
ing the domain and try to provide the reader an insight
about the ontology design requirements.
Cervical cancer is the second leading cause of
cancer-related deaths after breast cancer for women
between 20 and 39 years old (Landis et al., 1999) and
one of the leading types of cancer affecting women
worldwide. Despite a significant progress in early
diagnosis and treatment of cervical cancer, there are
more than 60,000 new cases and 30,000 deaths each
year in Europe alone. Recently, it has been proved
that infection by the human papilloma virus (HPV)
is necessary condition for the disease (Walboomers
et al., 1999). However, since HPV infection is highly
unlikely to be the sole cause for developingcancer, re-
cent trends in medical research combine genetic with
clinical data and attempt to discover underlying as-
sociations of the disease with environmental agents,
virus characteristics and genetic attributes, in order to
identify new markers of risk, diagnosis and prognosis.
Diagnosis of CxCa is based mainly on three types
of examinations, namely cytology, colposcopy, and
histology. A problem that arises here is that examina-
tions results are often encoded in Hospital Informa-
tion Systems (HIS) using different coding standards,
and/or custom formats.
On these grounds, a unification procedure is es-
sential. For medical research purposes (ie, for con-
ducting association studies), four levels of discretiza-
tion have been considered adequate for each exami-
nation (Agorastos et al., 2009). These are presented
in table 1.
Furthermore, in order to combine the findings of
these types of examinations, the severity of the dis-
ease is again quantized into four discrete levels, a
number that was considered adequate to capture it’s
different stages (Agorastos et al., 2009). When the
aggregate result needs to be based on more than one
types of examinations, the following practice is as-
sumed. If a histology has been conducted, then it’s re-
sult is considered regardless of the existence of other
results. In other words, histology is regarded the
”golden standard” examination having the highest re-
liability. Then follow colposcopy, and, finally, cytol-
ogy.
Another important aspect of the clinical research
procedure is that when more than one examinations of
the same type have been conducted, the ”worst” (ie,
most severe) is considered valid.
The same thing holds when a patient is associated
with more than one cases (ie, various series of exami-
nation and treatment procedures at different moments
in the course of time). Again, for this person, only
KEOD 2009 - International Conference on Knowledge Engineering and Ontology Development
104

Table 1: Summary of the adopted classification scheme for CxCa stages.
Normal (+Within Normal Limits) stage 0
Low Grade Cervical Intraepithelial Neoplasia (LCIN) stage 1
High Grade Cervical Intraepithelial Neoplasia (HCIN) stage 2
Invasive Cervical Cancer stage 3
it’s ”worst” case is taken under consideration for the
execution of an association study.
4 ONTOLOGY STRUCTURE
The conceptual schema of CxCa Ontology basically
consists of two types of entities (concepts).
1. Patient Management Entities. These are generic
entities capturing information about patient
records as they are stored in the corresponding
Hospital Information Systems (HIS). Their orga-
nization is based on the concepts of Case and
Visit.
A Case can be defined as ”a collection of data re-
ferring to a patient for a certain period of time,
within which a diagnosis on the disease is mean-
ingful, i.e., makes medical sense” and is related
to a Person. Each Case comprises of one or more
Visits. Every Visit is essentially a medical record
and may contain one or more Medical Interven-
tions. Each Medical Intervention is associated
with a single Result. However, since within the
context of a Case multiple Medical Interventions
of the same type with conflicting Results may be
conducted, each Case is associated with one and
only one Collective Result for each type of Medi-
cal Intervention.
When a diagnosis is made (i.e. the Severity Index
of the disease is calculated) it refers to a single
Case, taking into consideration all the Visits and
interventions the latter may be associated with.
This means that each Person is possibly related
with more than one Cases with different Diag-
noses through the hasCase object property. How-
ever, since for the purpose of performing an asso-
ciation study only one Case per Person has to be
considered, the hasWorstCase object property as-
sociates each Person with its Case with the worst
Diagnosis. A new Case is instantiated should the
patient return after a long period of time, which
would yield any previous examinations obsolete.
This type of entities also includes notions such
as Clinic, Medical Intervention, Result, Lifestyle
Choice etc.
2. Domain Specific Entities. These model terms that
are strongly associated with the CxCa disease, it’s
stages and the related interventions and genotypic
and phenotypic factors. This type of entities also
contains concepts that do not directly correspond
to the information stored in the EHR and are es-
sentially inferred through a reasoning process ac-
cording to appropriate definition rules.
Examples of such entities include Colposcopy,
HPV Vaccination, MTHFR C667T Polymorphism,
Stage1 CxCa Severity Index etc.
The scheme adopted is designed to be generic enough
to be potentially able to incorporate information about
other types of disease as well. The key-concepts of it,
along with their interconnecting properties, are repre-
sented in Figure 2.
Furthermore, the notion that the stages of the dis-
ease and the risk factors under investigation are not
affected by interrelations between different patients,
motivated us to design the ontology so that the ABox
forms independentindividual groups. This means that
two Person entities are neither directly nor indirectly
connected to each other. In this way, reasoning can be
performed independently within each subgraph con-
cerning a person, making the reasoning process an
explicitly parallel task.
5 MEDICAL UNIFICATION
RULES
The unification tasks of the ontology can be roughly
divided into two categories. The first category trans-
lates results from various different coding schemes to
a common format, while the second reveals implicit
classification criteria.
5.1 Result Unification Rules
The ontology contains all major standards for each
diagnostic intervention. Their results are translated
to a 4-category classification scheme for each type of
examination. For instance, in cytology, the Munich,
Bethesda and Pap coding schemes are considered and
characterization of a Cytology Result as a Stage0 Cy-
tology Result is based on satisfaction of the restriction
posed in formula (1).
AN ONTOLOGY FOR SUPPORTING CLINICAL RESEARCH ON CERVICAL CANCER
105
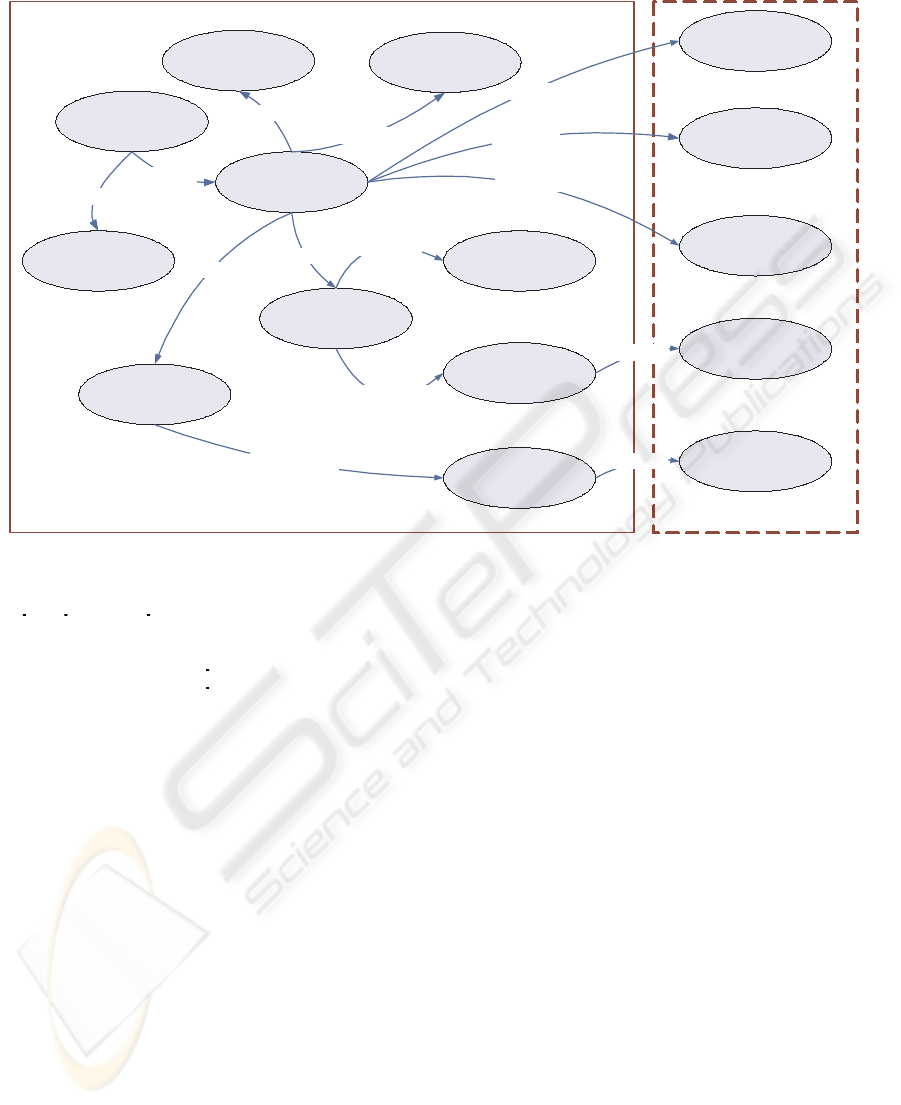
Person
Visit
HVP Test
Cytology
Histology
Case
Visit
hasCase
hasVisit
hasVisit
hasIntervention
hasIntervention
hasIntervention
Histology Result
CxCa Severity
Cytology Result
HPV Severity
hasResult
hasResult
hasSeverity
hasSeverity
Inferred Types
Smoking
Use of
Contraceptives
hasLifestyleChoice
hasLifestyleChoice
Polymorphism
hasPolymorphism
Smoking Severity
hasSeverity
Asserted Types
Figure 2: Basic Entities and their Interconnecting Properties.
stage0 Cytology Result ≡ Cytology Result ⊓
(∃isResultOfExam
(Cytology ⊓
((∃hasCytologyPapanikolaouResult {Class
II}) ⊔
(∃hasCytologyPapanikolaouResult {Class
I}) ⊔
(∃hasCytologyMunichResult {I}) ⊔
(∃hasCytologyMunichResult {II})⊔
(∃hasCytologyBethesdaResult {negative}) ⊔
(∃hasCytologyBethesdaResult {trichomonasVaginalis}) ⊔
(∃hasCytologyBethesdaResult {fungalOrganismsMorphConsistentWithCandidaSpp}) ⊔
(∃hasCytologyBethesdaResult{shiftInFloraSuggestiveBacterialVaginosis}) ⊔
(∃hasCytologyBethesdaResult{bacteriaMorphConsistentWithActinomycesSpp}) ⊔
(∃hasCytologyBethesdaResult{cellularChangesConsistentWithHerpesSimplexVirus}) ⊔
(∃hasCytologyBethesdaResult{otherNonNeoplasticFindings}) ⊔
(∃hasCytologyBethesdaResult{reactiveCellularChangesAssocWithInflammation}) ⊔
(∃hasCytologyBethesdaResult{reactiveCellularChangesAssocWithRadiation}) ⊔
(∃hasCytologyBethesdaResult{reactiveCellularChangesAssocWithIUD}) ⊔
(∃hasCytologyBethesdaResult{glandularCellsPostHysterectomyOrTrachelectomy}) ⊔
(∃hasCytologyBethesdaResult{atrophy}) ⊔
(∃hasResultDataType{stage0})
)
)
) (1)
5.2 Diagnostic Unification Rules
This type of rules aims at producing aggregate selec-
tion criteria for patient record retrieval.
An indispensable selection criterion when per-
forming association studies is the Severity Index (i.e.
the stage) of the disease. As there exist three types of
diagnostic interventions that may be associated with a
Case in our domain, calculation of the severity of the
specific Case is based upon them, giving priority to
existence of a Histology, in absence of which, consid-
ering Colposcopy and if both of the former are absent,
Cytology is considered.
As Description Logics adhere to a strict open
world assumption, it is impossible to deduce the ab-
sence of an examination result, as knowledge is con-
sidered incomplete. We overcome this problem by in-
stantiating ”triplets” of results (one for each exami-
nation) and explicitly storing a ”NoResult” property
for the examinations that have not actually been con-
ducted. An example of such a rule is given in formula
(2).
6 INFERENCE
Medical inference is essentially based on incomplete
knowledge and is thus non-monotonic. Doctors sug-
gest a treatment conjecturing about the most prob-
able cause for some observed symptoms or exami-
nation results, disregarding theoretically possible but
unlikely alternative causes. Moreover, these results
may contradict one another.
KEOD 2009 - International Conference on Knowledge Engineering and Ontology Development
106
stage1 Severity ≡ Cervical Cancer Severity ⊓
(∃isDiagnosedBy
(Case ⊓
((∃
caseHasCollectiveResult{ stage1 Collective Histology Result}) ⊔
((∃
caseHasCollectiveResult{ NO Collective Histology Result}) ⊓
(∃
caseHasCollectiveResult{ stage1 Collective Colposcopy Result})) ⊔
((∃
caseHasCollectiveResult{ NO Collective Histology Result}) ⊓
(∃
caseHasCollectiveResult{ NO
Collective Colposcopy Result}) ⊓
(∃
caseHasCollectiveResult{ stage1 Collective Cytology Result}))
)
)
) (2)
This is also the case in our setting. Before con-
cluding to a triplet of results associated with a case in
order to apply the rules of section 5.2, one has to pro-
duce a Collective Result for each examination type,
taking into account all examinations of this type.
Because there may be an undefined number of
each one of them, setting an upper limit of instances,
creating all of them and applying the trick of ”no re-
sult” as in section 5.2 is not an option here. In order
to overcome this problem we choose to add an inter-
mediate external inference step and consider closed
world semantics by using appropriate queries in an
RDF query language. This procedural layer executes
sequentially a number of queries, implementing the
set difference operator, in order of decreasing sever-
ity.
The process for doing this for cytology is as follows:
• First we query for cases containing cytologies as-
sociated with a stage3 Cytology Result. Obvi-
ously, since this is the worst outcome, these are
considered the Collective Cytology Results for the
corresponding cases.
• Then we query for examinations associated with
a stage2 Cytology Result. This query also returns
the Cytologies of the previous query, which are
subtracted with a proper query, and the remain-
ing ones are considered as the Collective Cytol-
ogy Results for the corresponding cases.
• The steps are continued for the other two stages
and the process is repeated for all examinations.
We employ the same methodology in order to
identify the worst Case for each Person. Again, con-
secutive queries are employed, this time retrieving
Persons w.r.t. their Collective Severity (as calculated
with the rules of section 5.2), in order of decreasing
severity.
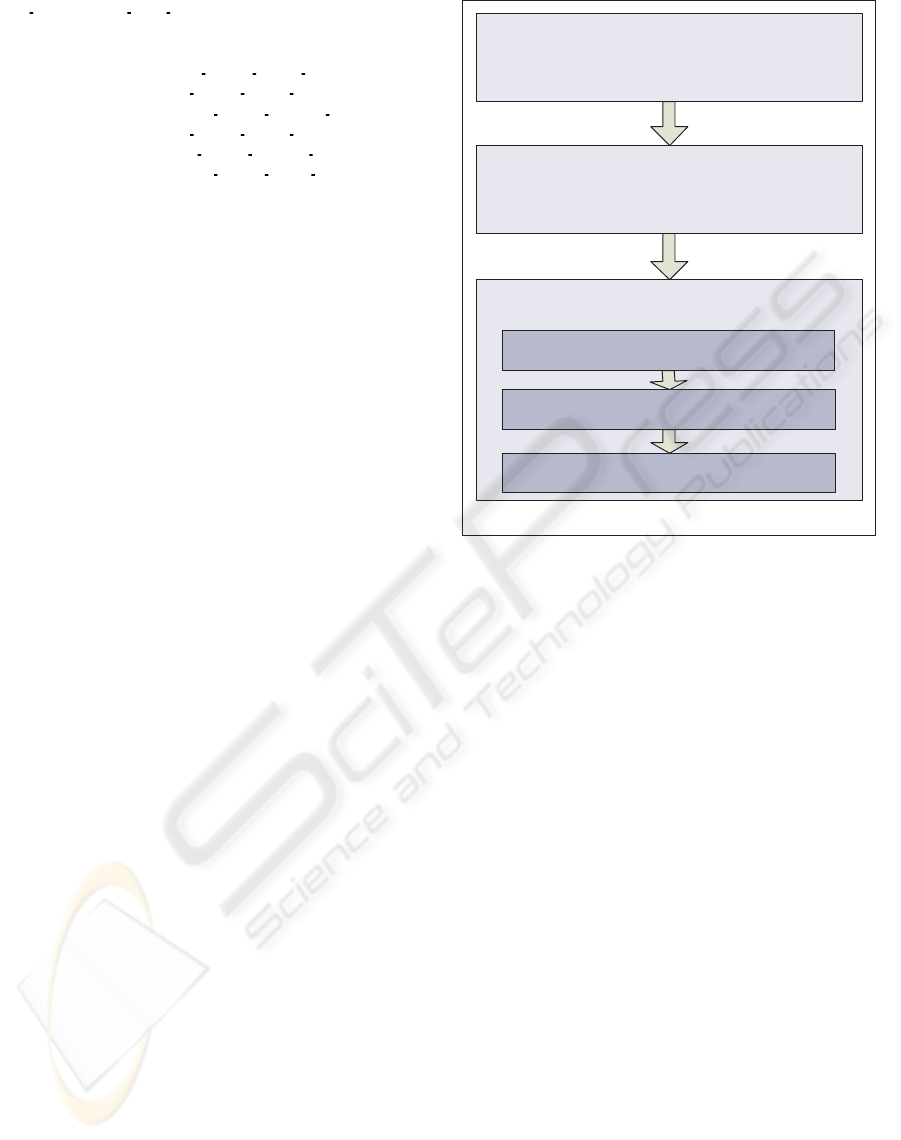
In summary, the unification procedure consists of
the steps depicted in Figure 3.
We have to point out that, because of it’s non-
monotonic nature, this process must be repeated each
time the ontology ABox is populated with new data.
Step 1:
Mapping to Ontology terms (Abox Instantiation)
Step 3
Calculation of Aggregate Metrics – Query Terms
Step 2:
Result Unification (through DL Reasoning)
Step 3.1:
Case-based Collective Result Calculation (Functional Programming)
Step 3.3:
Patient-based Worst Case Calculation (Functional Programming)
Step 3.2:
Case-based SI Calculation (through DL Reasoning)
Figure 3: The Unification Procedure.
However, this does not yield problems for our pur-
pose, as the system is research-oriented and does not
need to be concurrent w.r.t. the EHR records of the
associated hospitals.
7 IMPLEMENTATION DETAILS
The Ontology was developed in Protege knowledge
modeling tool (Noy et al., 2001), while the knowl-
edge representation language employed was OWL-
DL. Sesame (Broekstra et al., 2002) and OWLIM
(Kiryakov et al., 2005) were chosen for storage and
reasoning, respectively. DL reasoning is performed
by the IRRE component of Sesame and the queries
are expressed in SeRQL (Broekstra and Kampman,
2003).
The ontology TBox currently consists of 174
classes, 22 object and 96 datatype properties, 26
equivalent class axioms, while current instantiation of
the ontology ABox includes about 680,000 individual
620,000 role assertions, containing data about 3,200
patients and 8,400 cases.
AN ONTOLOGY FOR SUPPORTING CLINICAL RESEARCH ON CERVICAL CANCER
107

8 CONCLUSIONS
We have described the basic structure and inference
mechanisms of a medical ontology in the domain of
cervical cancer. The ontology is the central compo-
nent of a system that aims at the unification of various
virtual medical repositories and acts as a common lan-
guage designed as a means for conducting association
studies. Limitations and requirements of this cause
have made the use of an ad-hoc reasoning scheme in-
evitable.
ACKNOWLEDGEMENTS
This work was supported by the IST-2004-027510
project ‘Association Studies Assisted by Inference
and Semantic Technologies (ASSIST)’, funded by the
Commission of the European Community (CEC).
REFERENCES
Agorastos, T., Koutkias, V., Falelakis, M., Lekka, I., Mikos,
T., Delopoulos, A., Mitkas, P., Tantsis, A., Weyers,
S., Coorevits, P., Kaufmann, A., Kurzeja, R., and
Maglaveras, N. (2009). Semantic integration of cer-
vical cancer data repositories to facilitate multicenter
association studies: The assist approach. Cancer In-
formatics, 8:31–44.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D.,
Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K.,
Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P.,
Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese,
J. C., Richardson, J. E., Ringwald, M., Rubin, G. M.,
and Sherlock, G. (2000). Gene ontology: tool for
the unification of biology. The Gene Ontology Con-
sortium. Nature Genetics, 25(1):25–29.
Baader, F., Lutz, C., and Suntisrivaraporn, B. (2006). Cel - a
polynomial-time reasoner for life science ontologies.
In (Furbach and Shankar, 2006), pages 287–291.
Broekstra, J. and Kampman, A. (2003). The SeRQL Query
Language. Technical report, Aduna.
Broekstra, J., Kampman, A., and van Harmelen, F. (2002).
Sesame: A generic architecture for storing and query-
ing rdf and rdf schema. In Horrocks, I. and Hendler,
J. A., editors, International Semantic Web Conference,
volume 2342 of Lecture Notes in Computer Science,
pages 54–68. Springer.
Dyckhoff, R., editor (2000). Automated Reasoning with
Analytic Tableaux and Related Methods, International
Conference, TABLEAUX 2000, St Andrews, Scotland,
UK, July 3-7, 2000, Proceedings, volume 1847 of Lec-
ture Notes in Computer Science. Springer.
Furbach, U. and Shankar, N., editors (2006). Automated
Reasoning, Third International Joint Conference, IJ-
CAR 2006, Seattle, WA, USA, August 17-20, 2006,
Proceedings, volume 4130 of Lecture Notes in Com-
puter Science. Springer.
Hirschhorn, J. N. and Daly, M. J. (2005). Genome-wide
association studies for common diseases and complex
traits. Nature Reviews Genetics, 6(2):95–108.
Hustadt, U., Motik, B., and Sattler, U. (2004). Reduc-
ing shiq-description logic to disjunctive datalog pro-
grams. In Dubois, D., Welty, C. A., and Williams,
M.-A., editors, KR, pages 152–162. AAAI Press.
Jeen, P. H., Haase, P., Broekstra, J., Eberhart, A., and Volz,
R. (2004). A comparison of rdf query languages.
pages 502–517.
Kiryakov, A., Ognyanov, D., and Manov, D. (2005). Owlim
- a pragmatic semantic repository for owl. In Dean,
M., Guo, Y., Jun, W., Kaschek, R., Krishnaswamy, S.,
Pan, Z., and Sheng, Q. Z., editors, WISE Workshops,
volume 3807 of Lecture Notes in Computer Science,
pages 182–192. Springer.
Landis, S. H., Murray, T., Bolden, S., and Wingo, P. A.
(1999). Cancer statistics, 1999. CA Cancer J Clin.,
49(1):8–31.
Mitkas, P., Koutkias, V., Symeonidis, A., Falelakis, M.,
Diou, C., Lekka, I., Delopoulos, A., Agorastos, T.,
and Maglaveras, N. (2008). Association studies on
cervical cancer facilitated by inference and semantic
technologies: The assist approach. In International
Congress of the European Federation for Medical In-
formatics (MIE08), G¨oteborg, Sweden.
Noy, N. F., Sintek, M., Decker, S., Crub´ezy, M., Fergerson,
R. W., and Musen, M. A. (2001). Creating semantic
web contents with protege. IEEE Intelligent Systems,
16(2):60–71.
Sirin, E., Parsia, B., Grau, B. C., Kalyanpur, A., and Katz,
Y. (2007). Pellet: A practical owl-dl reasoner. J. Web
Sem., 5(2):51–53.
Tsarkov, D. and Horrocks, I. (2006). Description logic rea-
soner: System description. In (Furbach and Shankar,
2006), pages 292–297.
Walboomers, J. M., Jacobs, M. V., Manos, M. M., Bosch,
F. X., Kummer, J. A., Shah, K. V., Snijders, P. J.,
Peto, J., Meijer, C. J., and Munoz, N. (1999). Hu-
man papillomavirus is a necessary cause of invasive
cervical cancer worldwide. The Journal of Pathology,
189(1):12–19.
KEOD 2009 - International Conference on Knowledge Engineering and Ontology Development
108
