
DOCUMENT RETRIEVAL USING A PROBABILISTIC
KNOWLEDGE MODEL
Shuguang Wang
Intelligent Systems Program, University of Pittsburgh, Pittsburgh, PA, U.S.A.
Shyam Visweswaran
Department of Biomedical Informatics, University of Pittsburgh, Pittsburgh, PA, U.S.A.
Milos Hauskrecht
Department of Computer Science, University of Pittsburgh, Pittsburgh, PA, U.S.A.
Keywords:
Information retrieval, Link analysis, Domain knowledge, Biomedical documents, Probabilistic model.
Abstract:
We are interested in enhancing information retrieval methods by incorporating domain knowledge. In this pa-
per, we present a new document retrieval framework that learns a probabilistic knowledge model and exploits
this model to improve document retrieval. The knowledge model is represented by a network of associa-
tions among concepts defining key domain entities and is extracted from a corpus of documents or from a
curated domain knowledge base. This knowledge model is then used to perform concept-related probabilistic
inferences using link analysis methods and applied to the task of document retrieval. We evaluate this new
framework on two biomedical datasets and show that this novel knowledge-based approach outperforms the
state-of-art Lemur/Indri document retrieval method.
1 INTRODUCTION
Due to the richness and complexity of scientific do-
mains today, published research documents may fea-
sibly mention only a fraction of knowledge of the do-
main. This is not a problem for human readers who
are armed with a general knowledge of the domain
and hence are able to overcome the missing link and
connect the information in the article to the overall
body of domain knowledge. Many existing search
and information-retrieval systems that operate by an-
alyzing and matching queries only to individual doc-
uments are very likely to miss these knowledge-based
connections. Hence, many documents that are ex-
tremely relevant to the query may not be returned by
the existing search systems.
Our goal was to study ways of injecting knowl-
edge into the information retrieval process in order
to find better, more relevant documents that do not
exactly match the search queries. We present a new
probabilistic knowledge model learned from the asso-
ciation network relating pairs of domain concepts. In
general, associations may stand for and abstract a va-
riety of relations among domain concepts. We believe
that association networks and patterns therein give
clues about mutual relevance of domain concepts.
Our hypothesis is that highly interconnected domain
concepts define semantically relevant groups, and
these patterns are useful in performing information-
retrieval inferences, such as connecting hidden and
explicitly mentioned domain concepts in the docu-
ment.
The analysis of network structures is typically
done using link analysis methods. We adopt PHITS
(Cohn and Chang, 2000) to analyze the mutual con-
nectivity of domain concepts in association networks
and derive a probabilistic model that reflects, if the
above hypothesis is correct, the mutual relevance
among domain concepts. Figure 1 illustrates the dis-
tinction between our model and existing related tech-
niques. The top layer in Figure 1 consists of a set
of documents, and the bottom layer corresponds to
knowledge and relations among domain concepts (or
terms). The two layers are interconnected; individual
26
Wang S., Visweswaran S. and Hauskrecht M. (2009).
DOCUMENT RETRIEVAL USING A PROBABILISTIC KNOWLEDGE MODEL.
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval, pages 26-33
DOI: 10.5220/0002293400260033
Copyright
c
SciTePress
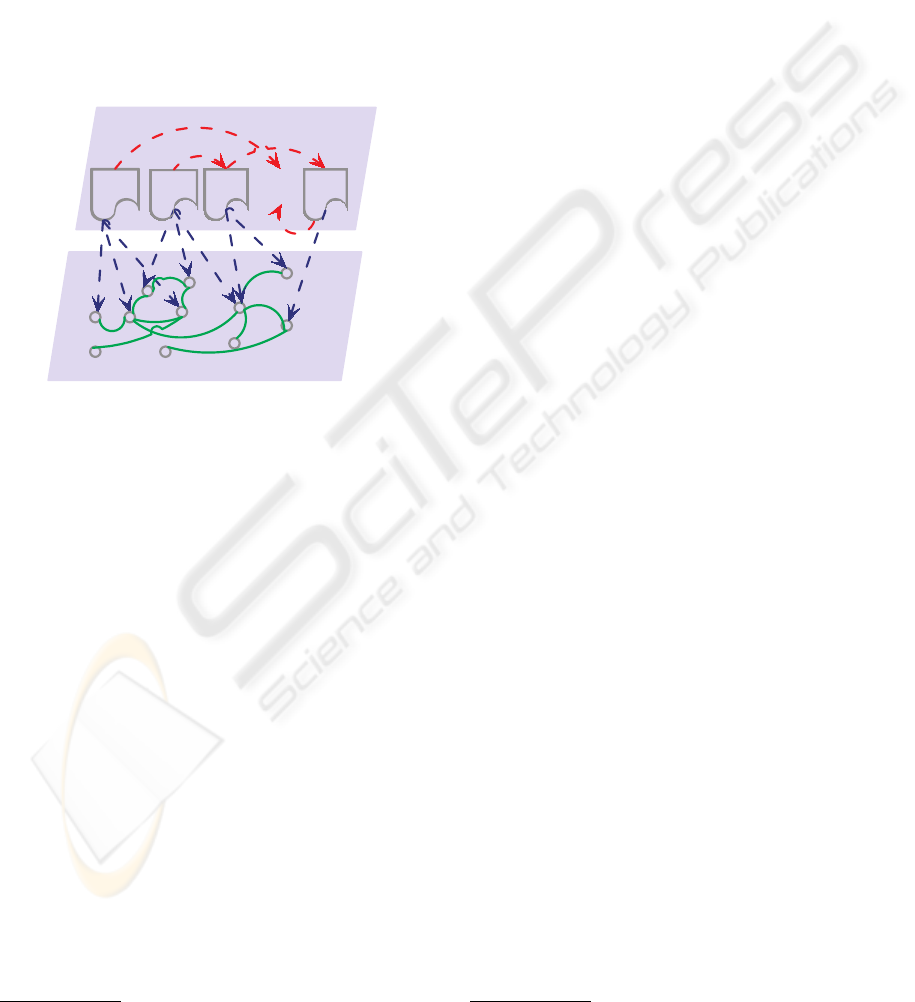
documents refer to multiple concepts and cite one an-
other. Latent semantic models, such as LSI (Landauer
et al., 1998) or PLSI (Hofmann, 1999), focus primar-
ily on document-term relations. Link analysis such
as PHITS is traditionally performed on the top (doc-
ument) layer and studies interconnections/citations
among documents to determine their dependencies.
In this work we use link analysis to study intercon-
nectedness of knowledge concepts (knowledge layer)
and their mutual influences. We believe that the
structure and interconnectedness in the knowledge
(and also on the document) layer has the potential to
greatly enhance standard information retrieval infer-
ences.
document
layer
knowledge
layer
doc1
...
doc2 doc3 docn
Figure 1: Illustration of the Knowledge Layer. Red dot-
ted arrows represent citations; blue dotted arrows link doc-
uments to the concepts mentioned in them; and green solid
lines connect associated concepts.
We experiment with and demonstrate the poten-
tial of our approach on biomedical research articles
using search queries on protein and gene species ref-
erenced in these articles. Our results show that the
addition of the knowledge layer inferences improves
the retrieval of relevant articles and outperforms the
state-of-the-art information retrieval systems, such as
the Lemur/Indri
1
.
This paper is organized as follows. Section 2
describes the knowledge model derived from con-
cept associations, and its construction from different
sources. Section 3 describe the learning of the proba-
bilistic knowledge model using PHITS and proposes
two inference methods to support document retrieval.
An extensive set of the experimental evaluations of
these methods is presented in Section 4. Several re-
cent related projects are reviewed in Section 5 and in
the final section we provide conclusions and some di-
rections for future work.
1
http://www.lemurproject.org/
2 THE KNOWLEDGE MODEL
The knowledge in any scientific domain can be rep-
resented as a rich network of relations among the do-
main concepts. Our information retrieval framework
adopts this kind of knowledge model to aid in the in-
formation retrieval process. The knowledge model
is represented by a graph (network) structure, where
nodes represent domain concepts and arcs between
nodes represent the pairwise relations among domain
concepts. In this paper, we focus on association rela-
tions, that abstract a variety of relations that may exist
among domain concepts.
Typically, a knowledge model is constructed by a
human expert. An example of such an expert-built
domain knowledge model is the gene and protein in-
teraction network that is available from the Molecu-
lar Signatures Database (MSigDB)
2
. Alternatively, a
knowledge model can be extracted from existing doc-
ument collections by mining and aggregating domain
concepts and their relations across many documents.
In the work described in this paper, we exper-
iment with knowledge models from the biomedical
domain with genes and proteins as domain concepts.
We analyze the utility of knowledge models extracted
from (1) the curated MSigDB database and (2) do-
main document collections. Knowledge extraction
from MSigDB is done in a simple fashion. Concepts
are already defined and the associations are based on
the grouping of different domain concepts, i.e., con-
cepts in the same group in the database are considered
to be associated. Knowledge extraction from the doc-
uments is done in two steps. In the first step, key do-
main concepts are identified using a dictionary look
up approach that will be presented in next section.
In the second step, relations (associations) are iden-
tified by analyzing the co-occurrence of pairs of do-
main concepts in the documents.
Domain concepts considered in our analysis con-
sist of genes and proteins. There are several freely
available resources that can be used to identify the
names of genes and proteins in text. However, these
resources are usually optimized with respect to F1
scores. To avoid introducing many false concepts and
relations into the model, we developed a method that
is optimized to achieve high precision. Briefly, our
method performs the following steps:
1. Segments abstracts (with titles) into sentences.
2. Tags sentences with MedPost POS tagger from
NCBI.
3. Parses tagged sentences with Collins’ full parser
(Collins, 1999).
2
http://www.broad.mit.edu/gsea/msigdb/
DOCUMENT RETRIEVAL USING A PROBABILISTIC KNOWLEDGE MODEL
27

4. Matches the phrases with the concept names
based on the GPSDB (Pillet et al., 2005) vocab-
ulary.
5. Matches the synonyms of concepts and assigns
unique id to distinct concepts.
This method archived over 90% precision at approx-
imately 65% recall when applied to a 100-document
test set to extract genes and proteins.
Once the concepts in the documents are identified,
we mine the associations among the concepts by ana-
lyzing co-occurrences of the concepts on the sentence
level. Specifically, two concepts are associated and
linked in the knowledge model if they co-occur in the
same sentence.
3 PROBABILISTIC KNOWLEDGE
MODEL
Our goal is to use the knowledge model represented
as an association network model to support inferences
on relevance among concepts. We propose to do so
by analyzing the interconnectedness of concepts in
the association network. More specifically, our hy-
pothesis is that domain concepts are more likely to
be relevant to each other if they belong to the same,
well defined, and highly interconnected group of con-
cepts. The intuition behind our approach is that con-
cepts that are semantically interconnected in terms of
their roles or functions should be considered more rel-
evant to each other. And we expect these semanti-
cally distinct roles and functions to be embedded in
the documents and hence picked up and reflected in
our association network.
To explore and understand the interconnectedness
of domain concepts in the association network we em-
ploy link analysis and the PHITS model (Cohn and
Chang, 2000).
3.1 Probabilistic HITS
PHITS (Cohn and Chang, 2000) is a probabilistic link
analysis model that was used to study graphs of co-
citation networks or web hyperlink structures. How-
ever, in our work, we use it to study relations among
domain concepts (terms) and not documents. This is
an important distinction to stress, since the PHITS
model on the document level has been used to im-
prove search and information retrieval performance
as well (Cohn and Chang, 2000). The novelty of our
work is in the use PHITS to learn the relations among
concepts and the development of a new approximate
inference method to assess the mutual relevancy of
domain concepts.
d
z
c
P(d|z)
P(z)
P(c|z)
d
z
c
P(d)
P(z|d)
P(c|z)
(a) Asymmetric parameterizations (b) Symmetric param eterizations
Figure 2: Graphical representation of PHITS.
Figure 2 shows a graphical representation of
PHITS. Variable d represents documents, z is the la-
tent factor, and c is a citation. There are two equiva-
lent PHITS parameterizations. Typically, a symmet-
ric parametrization is more efficient as the number of
topics z is smaller than the number of documents d.
Using the symmetric parametrization, the model de-
fines P(d,c) as
∑
z
P(z)P(c|z)P(z|d).
The parameters of the PHITS model are learned
from the link structure data using the Expectation-
Maximization (EM) approach (Hofmann, 1999; Cohn
and Chang, 2000). In the expectation step, it com-
putes P(z|d,c) and in the maximization step it re-
estimates P(z), P(d|z), and P(c|z).
The PHITS model has two important features.
First, PHITS (much like the PLSI model) is not a
proper generative probabilistic model for documents.
Latent Dirichlet Allocation (LDA) (Blei et al., 2003)
fixes the problem by using a Dirichlet prior to de-
fine the latent factor distribution. Second, the PHITS
model does not represent individual citations with
multiple random variables, instead citations are linked
to topics using a multinomial distribution over cita-
tions. Hence citations are treated as alternatives.
In our work, we use PHITS to analyze relations
among concepts. Hence both d (documents) and c
(citations) are substituted with domain concepts. To
make this difference clear we denote domain concepts
by e.
3.2 Document Retrieval Inferences with
PHITS
Our information retrieval framework assumes that
both documents and queries are represented by vec-
tors of domain concepts. Since individual research
articles usually refer only to a subset of domain con-
cepts a perfect match between the queries and the doc-
uments may not exist. Our goal is to develop methods
that use a knowledge model to infer absent but rele-
vant domain concepts.
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
28

We propose two approaches that use PHITS to im-
prove document retrieval. The first approach works
by expanding the document vector with relevant con-
cepts first and by applying information retrieval tech-
niques to retrieve documents afterwards. This ap-
proach can be easily incorporated into vector space
retrieval models. The second approach expands the
original query vectors with relevant concepts before
retrieving documents. This approach can be used in
most of information retrieval systems.
In the following sections, we first describe the ba-
sic inference supported by our model. After that we
show how this inference is applied in two retrieval ap-
proaches.
3.2.1 PHITS Inference
The basic inference task we support with the PHITS
model is the calculation of the probability of seeing an
absent (unobserved)concept e given a list of observed
concepts o
1
,o
2
,...,o
k
. As noted earlier, PHITS treats
concepts as alternatives and the conditional probabil-
ity is defined by the following distribution:
P(e = b
1
|o
1
,o
2
,...,o
k
)
P(e = b
2
|o
1
,o
2
,...,o
k
)
...
P(e = b
n
|o
1
,o
2
,...,o
k
),
where e is a random variable and b
1
,b
2
,...,b
n
are its
values that denote individual domain concepts. Intu-
itively, the conditional distribution defines the prob-
ability of seeing an absent concept next after we ob-
serve concepts in the document.
To calculate the conditional probability of e, we
use the following approximation:
P(e|o
1
,o
2
,...,o
k
,M
phits
)
=
∑
z
P(e|z,M
phits
)P(z|o
1
,o
2
,...,o
k
,,M
phits
)
∼
∑
z
P(e|z,M
phits
)
k
∏
j=1
P(z|o
j
,M
phits
) (1)
where o
1
,o
2
,...,o
k
are observed (known) concepts
and M
phits
is the PHITS model. We take an approx-
imation in this derivation due to the feature we dis-
cussed in Section 3.1 that it does not represent indi-
vidual citations with multiple random variables.
3.2.2 Document Expansion Retrieval
In information retrieval models, documents and
queries are usually represented as term vectors. Doc-
uments are retrieved according to a certain similar-
ity measure between documents and queries. In our
work, we use binary vectors to represent documents.
Explicitly mentioned concepts are represented as “1”s
in these vectors, and the others are “0”s initially.
However, concepts not explicitly mentioned in the
document may still be relevant to the document.
Our aim is to find a way to infer the probable rele-
vance of those absent concepts to the documents. Es-
sentially, these probable relevancevalues lets us trans-
form the original term vector for the document into a
new term vector, such that indicators of the terms in
the document are kept intact and terms not explicitly
mentioned in the document and their relevancies are
inferred as P(e
i
= T|d), where e
i
is an absent con-
cept in document d. Figure 3 illustrates the approach
and contrasts it with the typical information retrieval
process.
term vector
PLSA, LDA,
BM25 ...
1,x,x,1,x,x,x
1,0,0,1,0,0,0
1, 0.2 ,0.1 ,1, 0.05 ,0,0
Text DB
IR Methods
+ Query
IR Methods
+ Query
Curated KBPHITS KB
Figure 3: Exploitation of the domain knowledge model in
information retrieval. The standard method in which the
term vector is an indicator vector that reflects the occurrence
of terms in the document and query is on the left. Here
all unobserved terms are treated as zeros. In contrast, our
approach uses the knowledge model to fill in the values of
unobserved terms with their probabilities.
PHITS and inferences in Equation 1 treat concepts
as alternativesand the variable e ranges over all possi-
ble concepts. Hence in order to define P(e
i
|d), where
e
i
(different from e) is a boolean random variable that
reflects the probability of a concept i given the doc-
ument (or concepts explicitly observed in the docu-
ment) we need an approximation. We define the prob-
ability P(e
i
= T|d), with which a concept e
i
is ex-
pected or not expected to occur in the document d as:
P(e
i
= T|d) = min[α∗ P(e = b
i
|d), 1] (2)
where P(e = b
i
|d) is calculated from the PHITS
model using Equation 1 and α is a constant that scales
P(e = b
i
|d) to a new probability space. Constant α
can be defined in various ways. In our experiments
we assume α to be:
α = 1/min
j
P(e = b
j
|d)
where j ranges over all concepts explicitly mentioned
in the document d. An intuitive reason of this choice
DOCUMENT RETRIEVAL USING A PROBABILISTIC KNOWLEDGE MODEL
29

of α is that we do not want any absent concept to out-
weight any present concepts.
We expect the above domain knowledge infer-
ences to be applied before standard information re-
trieval methods are deployed. The row vector at the
top of Figure 3 is an indicator-based term-vector. This
term vector is either transformed with the help of
the knowledge model (our model) or retained with-
out change (standard model). We use the knowl-
edge model to expand the term vectors for all unob-
served concepts with their probabilities inferred from
the PHITS models. A variety of existing information
retrieval methods (e.g. PLSI (Hofmann, 1999), and
LDA (Wei and Croft, 2006)) can then be applied to
these two vector-term options. This makes it possible
to combine our knowledge model with many existing
retrieval techniques easily.
3.2.3 Query Expansion Retrieval
An alternative to expanding document vectors, is to
expand the query vectors. Briefly, the aim is to se-
lect concepts that are not in the original query, but are
likely to provide a relevant match. To implement this
approach, we first calculate P(e = b
i
|o
1
,o
2
,...,o
k
), the
probability of seeing an absent concept i given a list of
observed concepts o
1
,o
2
,...,o
k
in a query, as defined
in Equation 1. Then we sort the absent concepts ac-
cording to their conditional probabilities, and choose
the top m concepts to expand the query vector.
We applied two different inferences to expand
documents and queries because there is much less in-
formation or evidence in queries than that in docu-
ments. By choosing the top m relevant concepts we
can control the amount of noise introduced in the ex-
pansion. Our evaluation results show this approach is
effective to improve retrieval performance.
4 EXPERIMENTS
To demonstrate the benefit of the knowledge model,
we incorporate it in several document retrieval tech-
niques and compare them to the state-of-art research
search engine, Lemur/Indri. We learned the proba-
bilistic PHITS knowledge model from two sources:
the document corpus and the MSigDB database. Fur-
thermore, we combine these two knowledge models
using model averaging. We re-write Equation 1 as
follows to combine the inferences from the two mod-
els for model averaging:
P(e|d) =
∑
m
P(e|d, m)
P(d|m)P(m)
∑
m
P(d|m)P(m)
(3)
where e is an absent concept in document d and m is
a knowledge model. We assume uniform prior proba-
bility over the two models. Similarly, we apply model
averaging in all inference steps to combine the two
models.
We use the standard retrieval evaluation metric,
Mean Average Precision (MAP), to measure the re-
trieval performance in our experiments.
4.1 Document Expansion Retrieval
In the first set of experiments, we evaluate the doc-
ument expansion retrieval approach on a PubMed-
Cancer literature database. It consists of over 6000
research articles on 10 common cancers. Our corpus
contains both full documents and their abstracts.
Since document expansion can be easily incorpo-
rated into vector space information retrieval methods,
we combine it with two IR methods, namely, LDA
and PLSI. We compare it to Lemur/Indri with default
settings except that we use the Dirichlet smoothing
(µ = 2500), which had the best performance in our
experiments.
The knowledge model was learned from abstracts
and the complete text of the documents were used to
assess the relevance of documents returned by the sys-
tem. We randomly selected a subset of 20% of the ar-
ticles as the test set. To learn the probabilistic PHITS
knowledge model, we used i) 80% of the document
corpus, and ii) the MSigDB database. The combina-
tion of the two models was done at the inference level.
The relevance of a scientific document to the
query, especially if partial matches are to be assessed,
is best done by human experts. Unfortunately, this
is a very time-consuming process. So, we adopted
the following experimental setup: we perform all
knowledge-model learning and retrieval analysis on
abstracts only, and use exact matches of queries on
full texts as surrogate measures of true relevance.
Briefly, for a given query we retrieve a document
based on its abstract, and its relevance is judged (auto-
matically) by the query’s match to the full document.
We generated a set of 500 queries that consisted of
pairs of two domain concepts (proteins or genes) such
that 100 of these queries were generated by randomly
pairing any two concepts identified in the training cor-
pus, and 400 queries were generated using documents
in the test corpus by the following process. To gen-
erate a query we first randomly picked a test docu-
ment, and then randomly selected a pair of concepts
that were associated with each other in the full text of
this document. Thus, the generated query had a per-
fect match in the full text of at least one document.
All 500 queries were run on abstracts, and the rele-
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
30
vance of the retrieved document to the query was de-
termined by analyzing the full text and the match of
the query to the full text.
4.1.1 Document Retrieval Results
We applied the 500 queries to compare (1) PLSI and
LDA document retrievals with and without knowl-
edge expansion, and (2) document retrievals with
knowledge models mined from different sources to
the performance of the Lemur/Indri. Synonyms of
each query concept term are identified and appended
to the original query terms. To construct queries for
the Lemur/Indri, we use ‘boolean and’ to connect
the pair of query terms. An example of a query is:
“#band( #syn(synonyms of 1st gene) #syn(synonyms
of 2nd gene))”. Thus, using (1) we investigate if
probabilistic knowledge models help in improving re-
trieval performance and using (2) we compare the
utility of several knowledge sources for constructing
the knowledge model.
Table 1 gives the MAP scores of obtained by the
above mentioned methods utilizing knowledge mod-
els extracted from several sources. The different
knowledge sources are denoted by subscripts. ‘Tex-
tKM’ refers to methods that incorporate associations
from the document corpus; ‘MSigDBKM’ refers to
methods that include associations from the MSigDB
database; and ‘Text-MSigDBKM’ refers to methods
that include associations from both the corpus re-
fer to the relative improvement of the corresponding
method over the baseline Lemur/Indri method.
Overall, Lemur/Indri that ran on abstracts per-
formed significantly better than both the original
method, i.e., LDA and PLSI. However, all method
that incorporated a knowledge model performed bet-
ter than Lemur/Indri. Furthermore, combined ap-
proaches performed significantly better than the cor-
responding original approaches. These results show
that domain knowledge improves document retrieval
and supports our hypothesis that relevance is (at least
partly) determined by connectivity among domain
concepts. And it also confirms that domain knowl-
edge is helpful in finding relevant domain concepts.
Knowledge models that are mined from differ-
ent sources benefit the retrieval results differently.
Models mined from MSigDB did not help retrieval
as much as those mined from the document cor-
pus. This is partially due to its relatively small size:
the database contains about 3,000 unique genes in
over 300 groups. This also explains why combin-
ing knowledge from the document corpus and the
MSigDB database did not show a significant improve-
ment over knowledge obtained solely from corpus.
With a larger curated database, and thus a potentially
better knowledge model, we expect a larger improve-
ment in retrieval performance.
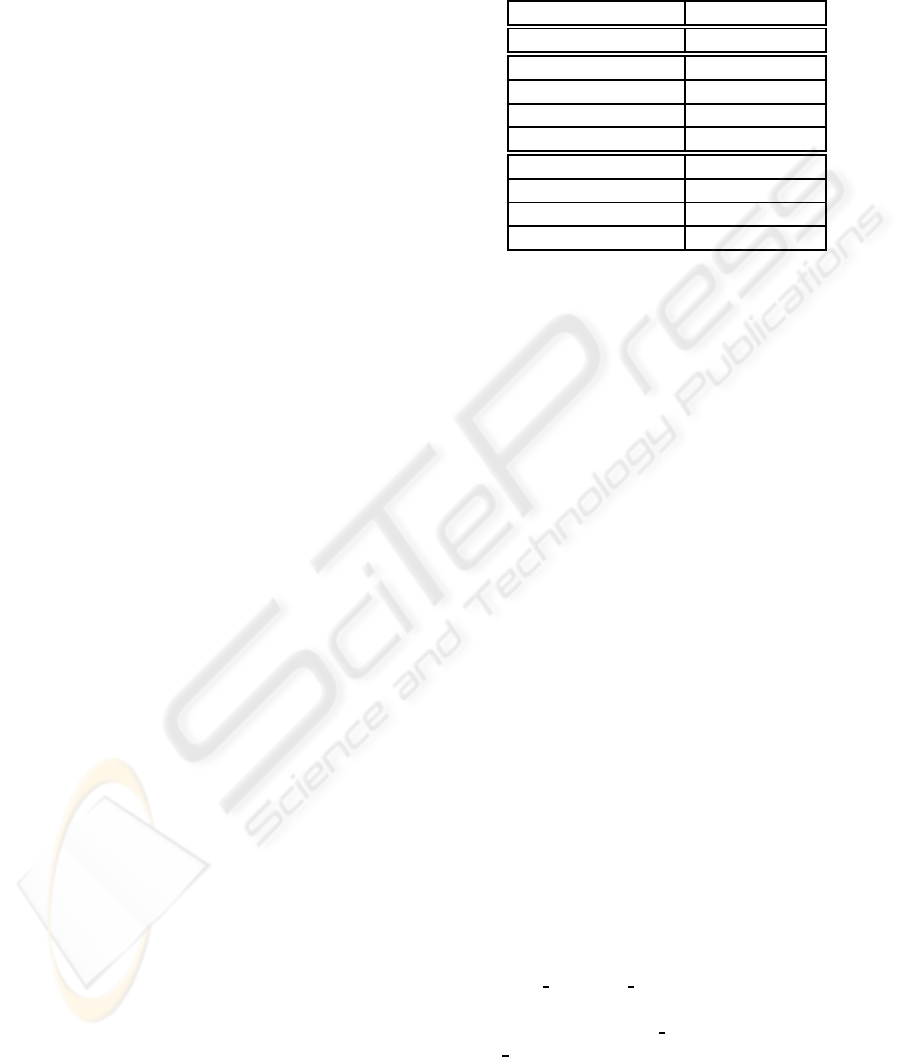
Table 1: MAP scores on PubMed-Cancer data.
Approaches MAP
Lemur/Indri 0.1891
PLSI 0.1633
PLSI
TextKM
0.1997 (+7%)
PLSI
MSigDBKM
0.1925 (+2%)
PLSI
Text+MSigDBKM
0.2001 (+7%)
LDA 0.1668
LDA
TextKM
0.2005 (+7%)
LDA
MSigDBKM
0.1977 (+5%)
LDA
Text−MSigDBKM
0.2009 (+7%)
4.2 Query Expansion Retrieval
In the second set of experiments, we compare the
query expansion approach with the pseudo-relevance
feedback expansion model in Lemur/Indri. In addi-
tion to the PubMed-Cancer dataset, these experiments
included the TREC Genomic Track 2003 dataset.
The Genomic Track dataset is significantly larger
than the Pubmed-Cancer dataset and consists of over
520,000 abstracts from Medline. The dataset comes
with a set of test queries used for the evaluation of
retrieval methods. Evaluation queries consist of gene
names and their aliases, with the specific task being
derivedfrom the definition of GeneRIF. The relevance
of the documents to test queries for the TREC data
were assessed by human experts.
Although queries contain only gene names, the
dataset itself consists of a wider range of biomedical
topics. Again, we learn the knowledge models from
two sources. To learn the knowledge model from the
TREC dataset, we used only 1/3 of the dataset for the
sake of efficiency.
We expanded the original queries with 10 terms
(m=10) from i) an internal query expansion module
in Lemur/Indri that is based on a pseudo-relevance
feedback model (Lavrenko and Croft, 2001), and ii)
with the most “relevant” concepts inferred from our
probabilistic knowledge models. The following query
provides an example:
original query
#combine(o concept1 o concept2 ...)
expanded query
#weight(2.0 #combine( o concept1 ...) 1.0 #com-
bine(e concept1 ...))
We use the higher (double) weights for the terms
in the original query compared to the expanded terms.
Table 2 gives the MAP scores of Lemur/Indri with
various settings on the PubMed-Cancer and Genomic
DOCUMENT RETRIEVAL USING A PROBABILISTIC KNOWLEDGE MODEL
31

Track datasets. We compare (1) different expan-
sion approaches, and (2) knowledge-drivenexpansion
with three different PHITS models (same as those
used for the experiments described in the previous
section). The relative percents in brackets represent
relative improvements of various methods over the
baseline Lemur/Indri. The query expansion methods
that use knowledge models (last three rows) show im-
proved retrieval performance. Furthermore, all these
methods outperform the pseudo-relevance feedback
model in Lemur/Indri.
On comparing the effect of knowledge models
mined from different sources, the models that include
associations from the document corpus perform better
than models extracted from the curated database. We
believe that this is again due to the relatively small
size of the database and the sparse association net-
work it induces.
On comparing the performance on two datasets,
the relative improvement is smaller for all query ex-
pansion approaches on the PubMed-Cancer data. This
retrieval task is more difficult since the queries that we
constructed are extracted from the full text of the doc-
uments, and hence many the of query terms may not
even appear in the abstracts. Thus, document expan-
sion is a better choice when documents contain more
evidence concepts than queries.
Table 2: MAP scores on TREC Genomic Track 03 and
PubMed-Cancer data.
Approaches TREC PubMed-Cancer
Lemur/Indri 0.2568 0.1891
+relevance 0.2643(+3%) 0.1948(+3%)
+TextKM 0.2773(+8%) 0.1983(+4%)
+MSigDBKM 0.2688(+5%) 0.1951(+3%)
+Text-MSigDBKM 0.2778(+8%) 0.1985(+4%)
5 RELATED WORK
In the context of information retrieval, domain knowl-
edge has been used in (Zhou et al., 2007) and (Pick-
ens and MacFarlane, 2006). (Pickens and MacFar-
lane, 2006) showed that the occurrences of terms can
be better weighted using contextual knowledge of the
terms. (Zhou et al., 2007) presented various ways
of incorporating existing domain knowledge from
MeSH and Entrez Gene and demonstrated improve-
ment in information retrieval. (B¨uttcher et al., 2004)
expanded the queries with synonyms of all biomed-
ical terms extracted from external databases. In ad-
dition to synonyms, (Aronson and Rindflesch, 1997)
mapped the terms in the queries to biomedical con-
cepts using MetaMap and added these concepts to the
original queries. (Lin and Demner-Fushman, 2006)
showed that it is beneficial to use a knowledge model.
In terms of knowledge extraction, (Lee et al., 2007)
presented a method for finding important associations
among GO and MeSH terms and for computing con-
fidence and support scores for them. All these ap-
proaches differ from our approach in that we extract
domain concepts and relations from a document cor-
pus. Then we learn a probabilistic knowledge model
from the association network automatically and ex-
ploit it to infer the missing knowledge in the individ-
ual documents.
6 CONCLUSIONS AND FUTURE
WORK
We have presented a new framework that extracts do-
main knowledge from multiple documents and a cu-
rated domain knowledge base and uses it to support
document retrieval inferences. We showed that our
method can improve the retrieval performance of doc-
uments when applied to the biomedical literature. To
the best of our knowledge this is the first study that at-
tempts to learn probabilistic relations among domain
concepts using link analysis methods and performs
inference in the knowledge model for document re-
trieval.
The inference potential of our framework in re-
trieval of relevant documents was demonstrated in the
experiments with document abstracts, in which full
documents and relations therein were used only to as-
sess quantitatively the relevance of the document to
the query. This result was confirmed by further ex-
periments on the Genomic track dataset.
Our knowledge model was extracted using asso-
ciations among domain-specific terms observed in a
document corpus and from curated knowledge col-
lected in a database. We did attempt to refine these
associations and identify the specific relations they
represent. However, we anticipate that the use of a
more comprehensive knowledge model with a vari-
ety of explicitly represented relations among the do-
main concepts will further improve the information
retrieval performance. At the same time, we are look-
ing into other inference alternatives to avoid taking
approximations. Our experiments show that knowl-
edge models mined from the literature perform better
in document retrieval. This is partially due to the dif-
ficulty in locating an appropriate knowledge base for
each retrieval task. Finally, our model is robust and
flexible enough to integrate knowledge from various
sources and combine them with existing document re-
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
32

trieval methods.
REFERENCES
Aronson, A. R. and Rindflesch, T. C. (1997). Query ex-
pansion using the umls metathesaurus. In TREC ’04:
Proceedings of the AMIA Annual Fall Symposium 97,
JAMIA Suppl, pages 485–489. AMIA.
Blei, D. M., Ng, A. Y., and Jordan, M. I. (2003). Latent
dirichlet allocation. The Journal of Machine Learning
Research, 3:993–1022.
B¨uttcher, S., Clarke, C. L. A., and Cormack, G. V. (2004).
Domain-specific synonym expansion and validation
for biomedical information retrieval. In TREC ’04:
Proceedings of the 13th Text REtrieval Conference.
Cohn, D. and Chang, H. (2000). Learning to probabilisti-
cally identify authoritative documents. In Proc. 17th
International Conf. on Machine Learning, pages 167–
174. Morgan Kaufmann, San Francisco, CA.
Collins, M. (1999). Head-driven statistical models for nat-
ural language parsing. PhD Dissertation.
Hofmann, T. (1999). Probabilistic latent semantic index-
ing. In SIGIR ’99: Proceedings of the 22nd annual
international ACM SIGIR conference on research and
development in information retrieval, pages 50–57.
ACM Press.
Landauer, T. K., Foltz, P. W., and Laham, D. (1998). In-
troduction to latent semantic analysis. Discourse Pro-
cesses, 25:259–284.
Lavrenko, V. and Croft, B. W. (2001). Relevance based
language models. In SIGIR ’01: Proceedings of the
24th annual international ACM SIGIR conference on
Research and development in information retrieval,
pages 120–127. ACM.
Lee, W.-J., Raschid, L., Srinivasan, P., Shah, N., Rubin,
D., and Noy, N. (2007). Using annotations from con-
trolled vocabularies to find meaningful associations.
In DILS ’07: In Fourth International Workshop on
Data Integration in the Life Sciences, pages 27–29.
Lin, J. and Demner-Fushman, D. (2006). The role of knowl-
edge in conceptual retrieval: a study in the domain of
clinical medicine. In SIGIR ’06: Proceedings of the
29th annual international ACM SIGIR conference on
Research and development in information retrieval,
pages 99–106. ACM.
Pickens, J. and MacFarlane, A. (2006). Term context mod-
els for information retrieval. In CIKM ’06: Proceed-
ings of the 15th ACM international conference on In-
formation and knowledge management, pages 559–
566. ACM.
Pillet, V., Zehnder, M., Seewald, A. K., Veuthey, A.-L., and
Petra, J. (2005). Gpsdb: a new database for synonyms
expansion of gene and protein names. Bioinformatics,
21(8):1743–1744.
Wei, X. and Croft, W. B. (2006). Lda-based document mod-
els for ad-hoc retrieval. In SIGIR ’06: Proceedings of
the 29th annual international ACM SIGIR conference
on research and development in information retrieval,
pages 178–185. ACM.
Zhou, W., Yu, C., Smalheiser, N., Torvik, V., and Hong,
J. (2007). Knowledge-intensive conceptual retrieval
and passage extraction of biomedical literature. In SI-
GIR ’07: Proceedings of the 30th annual international
ACM SIGIR conference on research and development
in information retrieval, pages 655–662. ACM.
DOCUMENT RETRIEVAL USING A PROBABILISTIC KNOWLEDGE MODEL
33
