
DILIA - A DIGITAL LIBRARY ASSISTANT
A New Approach to Information Discovery through Information Extraction
and Visualization
Inessa Seifert
1
, Kathrin Eichler
2
, Holmer Hemsen
2
, Sven Schmeier
2
, Michael Kruppa
1
Norbert Reithinger
1
and G
¨
unter Neumann
2
1
Intelligent User Interfaces,
2
Language Technology, DFKI (German Research Center for Artificial Intelligence)
Alt-Moabit 91 C, 10559 Berlin, Germany
Keywords:
Digital libraries, Technical term extraction, Information discovery, Visualization, Co-author networks.
Abstract:
This paper presents preliminary results of our current research project DiLiA (Digital Library Assistant).
The goals of the project are are twofold. One goal of the project is the development of domain-independent
information extraction methods. The other goal is the development of information visualization methods that
interactively support researchers at time consuming information discovery tasks. We first describe issues
that contribute to high cognitive load during exploration of unfamiliar research domains. Then we present a
domain-independent approach to technical term extraction from paper abstracts, describe the architecture of
the DiLiA, and illustrate an example co-author network visualization.
1 INTRODUCTION
This paper presents preliminary results of our current
research project DiLiA (Digital Library Assistant).
Our research goals are twofold. One goal of the
project is the development of sophisticated domain-
independent information extraction techniques that
aim at retrieving specific entities (e.g., technical
terms, key ideas) and relations (e.g., citations, co-
authorships) among the documents contained in a dig-
ital library.
The other goal of the project involves the develop-
ment of sophisticated visualization methods in order
to interactively support researchers at time consuming
information seeking tasks. These methods should vi-
sually present huge result sets caused by vaguely de-
fined search queries and allow the information seek-
ers to examine, analyze, and manipulate multitudi-
nous dimensions of query results from various per-
spectives.
Finally, we aim at combining these two techniques
to make the extracted structures and relations con-
cealed in result sets transparent to information seek-
ers.
In this paper, we will exemplify aspects that con-
tribute to the cognitive complexity of information dis-
covery tasks. We will outline information extraction
methods that can be used for pre-processing of data
contained in digital libraries. In doing so, parts of
the mental work that has to be accomplished by the
information seeker can be offloaded to the assisting
system. We will discuss characteristic requirements
for data visualization in digital libraries. Finally, we
will conclude with an example visualization that il-
lustrates our preliminary results.
2 INFORMATION SEEKING
Information seeking is a complex and cognitively de-
manding task that has a close relation to learning
and problem solving (Vakkari, 1999). The informa-
tion seeking process starts with an initial concept of
a search goal that is derived from the user’s prior
knowledge about the problem domain. Based on this
knowledge, the information seeker defines an initial
search query. The analysis of the retrieved query re-
sults contributes to generation of new concepts, revi-
sion of search goals, and formulation of new queries.
Concepts, search goals, as well as criteria for assess-
ing the relevance of articles from the query results
evolve during the information seeking process and
cannot be specified in advance (Bates, 1989).
The lack of domain specific knowledge leads to
180
Seifert I., Eichler K., Hemsen H., Schmeier S., Kruppa M., Reithinger N. and Neumann G. (2009).
DILIA - A DIGITAL LIBRARY ASSISTANT - A New Approach to Information Discovery through Information Extraction and Visualization.
In Proceedings of the International Conference on Knowledge Management and Information Sharing, pages 180-185
DOI: 10.5220/0002304001800185
Copyright
c
SciTePress

underdetermined and unclear search goals that are re-
flected in the definition of vague search queries. Such
search queries contribute to a huge number of result-
ing hits. Examining a great amount of scientific liter-
ature is a time consuming endeavor.
Each article is distinguished by a title, authors, a
short description (i.e., abstract), a source (e.g., book,
journal, etc.), publishing date (e.g., year), and its text.
These attributes can contain specific words, i.e., terms
that can be recognized by the information seeker as
relevant and trigger the formulation of refined search
queries (Barry, 1994; Anderson, 2006).
Studies conducted by (Anderson, 2006) reported
that it was difficult to find and specify appropriate
terms to define more precise search queries, espe-
cially, if an information seeker was unfamiliar with
the terminology of the problem domain, or if this ter-
minology changed over time.
3 INFORMATION EXTRACTION
Our idea for domain-independent term extraction is
based on the assumption that, regardless of the do-
main we are dealing with, the majority of the TTs in
a document are in nominal group positions. To ver-
ify this assumption, we manually annotated a set of
100 abstracts from the biology part of the Zeitschrift
fuer Naturforschung
1
(ZfN) archive, which contains
scientific papers published by the ZfN between 1997
and 2003. We found that 94% of the annotated terms
were in fact in noun group positions. The starting
point of our method for extracting terms is therefore
an algorithm to extract nominal groups from a text.
We then classify these nominal groups into TTs and
non-TTs using frequency counts retrieved from the
MSN search engine. For the extraction of term can-
didates, we use the nominal group (NG) chunker of
the GNR tool developed by (Spurk, 2006), which we
slightly adapted for our purposes. The advantage of
this chunker compared to other chunkers is that it is
domain-independent because it is not trained on a par-
ticular corpus but relies on patterns based on closed
class words (e.g. prepositions, determiners, coordi-
nators), which are available in all domains. Using
lists of closed-class words, the NG chunker deter-
mines the left and right boundaries of a word group
and defines all words in between as an NG. In order
to find the TTs within the extracted NG chunks, we
use a frequency-based approach. Our assumption is
that terms that occur mid-frequently in a large cor-
pus are the ones that are most associated with some
1
http://www.znaturforsch.com/
topic and will often constitute technical terms. To
test our hypothesis, we retrieved frequency scores for
all NG chunks extracted from our corpus of abstracts
from the biology domain and calculated the ratio be-
tween TTs and non-TTs for particular maximum fre-
quency scores. To retrieve the frequency scores for
our chunks, we use the internet as reference corpus,
as it is general enough to cover a broad range of do-
mains, and retrieve the scores using the Live Search
API of the MSN search engine
2
. The results confirm
our hypothesis, showing that the ratio increases up to
an MSN score threshold of about 1.5 million and then
slowly declines. This means that chunks with mid-
frequency score are in fact more likely to be technical
terms than terms with a low or high score.
To optimize the lower and upper boundaries that de-
fine ’mid-frequency’, we maximized the F-measure
achieved on our annotated biology corpus with dif-
ferent thresholds set. Evaluating our algorithm on our
annotated corpus of abstracts, we obtained the follow-
ing results. From the biology corpus, our NG chunker
was able to extract 1264 (63.2%) of the 2001 anno-
tated TTs in NG position completely and 560 (28.0%)
partially. With the threshold optimized for the F-
measure (6.05 million), we achieved a precision of
57.0% at recall 82.9% of the total matches. These re-
sults are comparable to results for GN learning, e.g.
those by (Yangarber et al., 2002) for extracting dis-
eases from a medical corpus. We also evaluated our
approach on the GENIA corpus
3
, a standard corpus
for biology. Considering all GENIA terms with POS
tags matching the regular expression
JJ ∗ NN ∗ (NN|NNS)
as terms in NG position, we were able to evaluate our
approach on 62.4% of all terms. With this data, we
achieved 50.0% precision at recall 75.0%. A sample
abstract from the ZfN data, with the automatically ex-
tracted TTs shaded, is shown in Figure 1. The key
advantage of our approach over other approaches to
GN learning is that it extracts a broad range of differ-
ent TTs robustly and irrespective of the existence of
morphological or contextual patterns in a training cor-
pus. It works independent of the domain, the length of
the input text or the size of the corpus, in which in the
input document appears. This makes it, in principal,
applicable to documents of any digital library.
2
http://dev.live.com/livesearch/
3
http://www-tsujii.is.s.u-tokyo.ac.jp/genia/topics/ Cor-
pus/
DILIA - A DIGITAL LIBRARY ASSISTANT - A New Approach to Information Discovery through Information
Extraction and Visualization
181
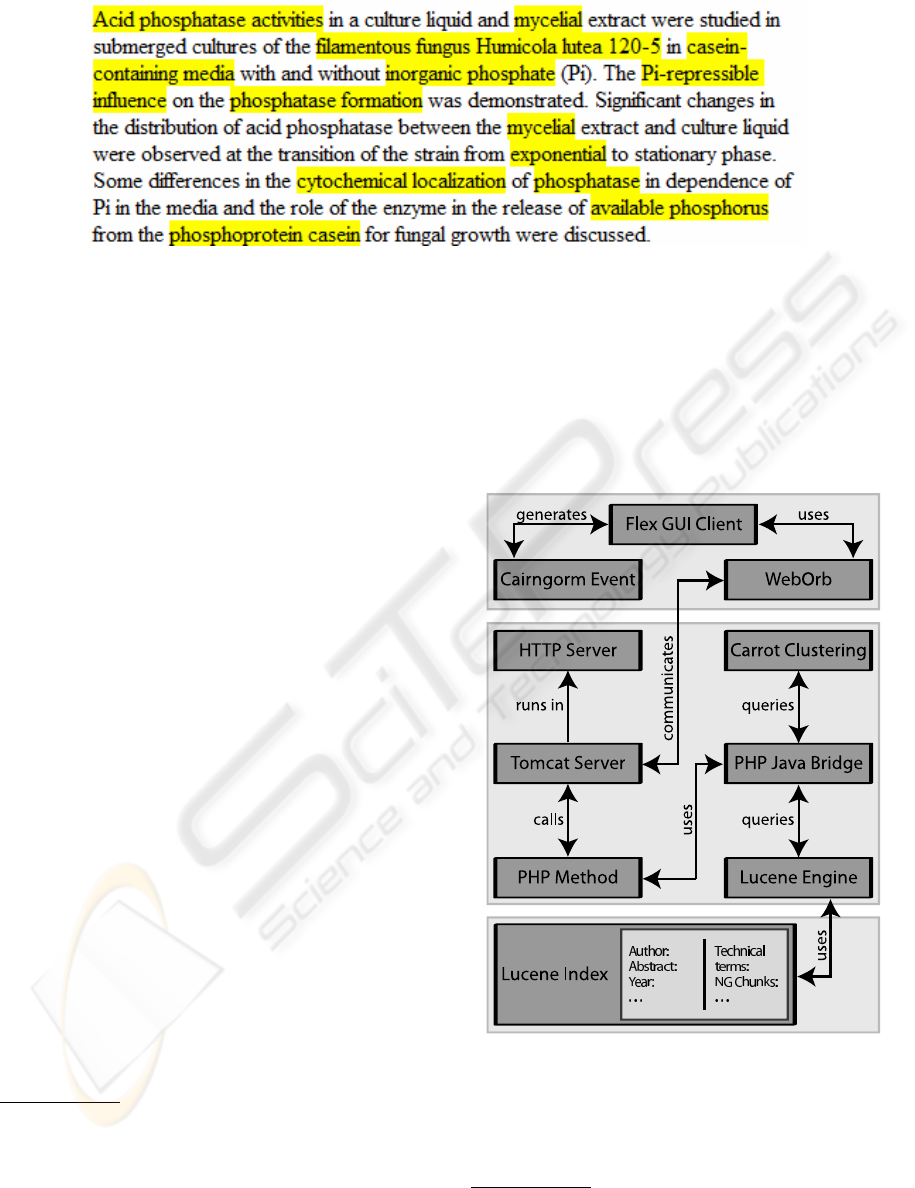
Figure 1: Sample output of our TT extraction algorithm.
4 ARCHITECTURE
The system’s architecture makes use of several stan-
dardized paradigms in order to guarantee a robust,
scalable application that is based on reusable com-
ponents. It consists of a 3-tier web-based client
server architecture. The client side has been devel-
oped as a Rich Internet Application (RIA) realized
in Adobe Flex
4
. This application follows the model-
view-controller (MVC) concept. The flex prototype
makes use of the Cairngorm
5
MVC implementation
which ensures a consequent MVC realization. The
client utilizes server side PHP
6
classes to query the
digital library database. The queries are executed
by the Lucene Search Engine
7
. The Lucene Index
holds all documents of the digital library including
their metatags like author, headline, abstract, publish-
ing year, etc.. The results of the described information
extraction (see section 3) are represented as additional
metatag fields of a document. As the metatag repre-
sentation in the index is realized by separate fields it is
possible to formulate search queries that search only
in a subset of all metatags. With this the impact of the
described information extraction results can be mea-
sured directly.
The connection between PHP and the Lucene
Search engine is established by the PHP Javabridge
8
. Finally, the communication between Flex (which
is compiled into a Flash Movie) and the server side
PHP classes is realized using Weborb
9
. Weborb han-
dles the serialization/deserialization of data and the
interfacing of methods between PHP and Flex. To de-
termine the topic labels, we use the Carrot clustering
4
http://www.adobe.com/products/flex/
5
http://opensource.adobe.com/wiki/display/cairngorm/
6
http://www.php.net
7
http://lucene.apache.org/
8
http://php-java-bridge.sourceforge.net
9
http://www.themidnightcoders.com/products/weborb-
for-php/
engine
10
which is fed with the results of the Lucene
Search Engine. Thus the results of the informa-
tion extraction also influence the clusters topics.The
server side environment is based on the Apache HTTP
Server
11
and Apache Tomcat
12
. The information flow
between server and client is visualized in Figure 2.
Figure 2: The general DILIA architecture.
10
http://project.carrot2.org/
11
http://httpd.apache.org/
12
http://tomcat.apache.org/
KMIS 2009 - International Conference on Knowledge Management and Information Sharing
182

5 INFORMATION
VISUALIZATION
The main purpose of the information visualization
techniques is to present the data contained in a dig-
ital library and provide interactive operations to the
information seeker that facilitate exploration of its
content. Commonly used visualizations include a
query panel for the formulation of search queries and
a simple hit list that presents meta information such
as author’s names, title, etc. (e.g., search engines
such as www.google.com). Digital libraries special-
ized in specific research fields provide a possibil-
ity for browsing in manually annotated categories,
journal, conference, or workshop catalogs that con-
vey an overview about a research topic and facilitate
the exploration (see e.g., http://www.lt-world.org/).
Recently developed domain-independent search en-
gines employ clustering algorithms that allow for ef-
ficient online-clustering of query results into topics
that can be used for filtering of information or further
query formulations (see, e.g., http://www.cuil.com/,
http://www.kartoo.com/, http://www.quintura.com/).
Alternatively to hit lists, digital libraries offer
graph-based representations of hierarchically struc-
tured topics, citation and co-author networks.
Spatially inspired concept spaces display differ-
ent concepts that involve central terms retrieved from
clustered query results (Zhang et al., 2002). Spatial
distance between the concepts conveys similarity re-
lations between the extracted terms.
Topic maps provided by HighWire digital library
consist of tree-based structures that include hierarchi-
cally structured topics and subtopics
13
. The interac-
tive operations allow for expanding topics in order to
reach a finer level of granularity.
3D-visualizations present the content of a dig-
ital library as cone trees (Robertson et al., 1991;
Mizukoshi et al., 2006). Cones stand for different top-
ics and subtopics that contain documents represented
as leaves of a tree. The user can interactively rotate
the cones to examine the titles of the documents.
The major problem in the visualization of cita-
tion and co-author networks is a great number of
documents and a high connectivity of scientific pa-
pers. Large graphs compromise the performance of
assisting systems and contribute to mental informa-
tion overload, since they are hard to understand (Her-
man et al., 2000). One of the approaches, for example,
reduces the amount of edges leading from one arti-
cle to another by employing a minimal tree-spanning
algorithm for extraction of shortest paths connecting
13
http://highwire.stanford.edu/help/hbt/
the articles (Elmqvist and Tsigas, 2007). Displaying
only these paths allowed for better visual inspection
of citation clusters. Although the works described so
far cover specific aspects of this problem, it is still an
open research question how to efficiently combine in-
formation extraction techniques with interactive visu-
alizations to support the information discovery during
exploration of scientific literature.
In the following section, we will present an ex-
ample visualization of a co-author network that can
be filtered according to the topics extracted from ab-
stracts of cooperatively published papers.
6 AN EXAMPLE CO-AUTHOR
NETWORK
We used the data of the DBLP Computer Science
Bibliography
14
to resolve the co-author relations be-
tween scientific publications. The DiLiA user inter-
face implements basic functionality that enables the
user to formulate a search query and receive a list of
publications as a result. The user can select either an
article or an author from the generated result list in
order to analyze the scientific cooperations in a co-
author network. The following figures illustrate two
different views on the co-author network of Andreas
Dengel who is a well known scientist in the knowl-
edge representation and management community.
Figure 3: An example co-author network of “Andreas Den-
gel”.
The first view (Fig. 3) presents the author in the
center, his publications in the first row, and corre-
sponding collaborators in the second row. Since this
14
http://www.informatik.uni-trier.de/ ley/db/
DILIA - A DIGITAL LIBRARY ASSISTANT - A New Approach to Information Discovery through Information
Extraction and Visualization
183
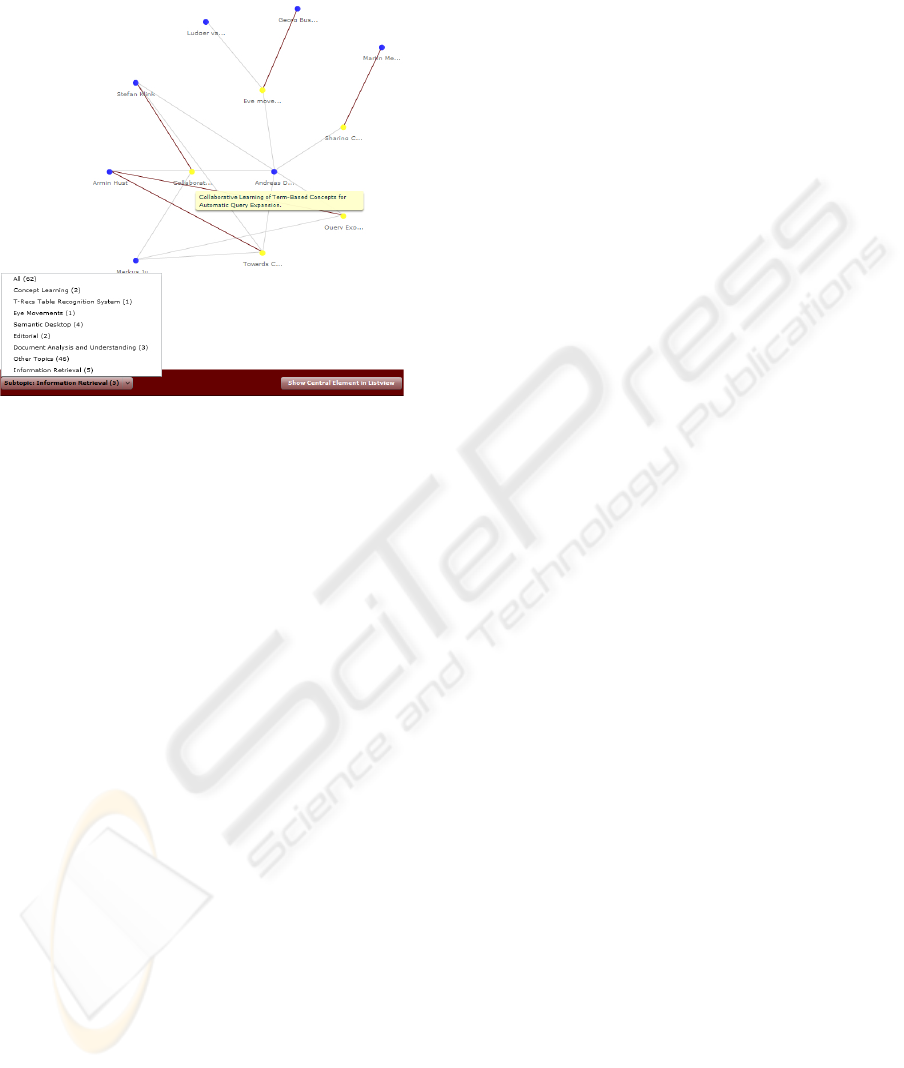
author published a lot of papers in his scientific career,
the co-author graph is considerably large.
Figure 4: Filtering the co-author network of “Andreas Den-
gel” according to the topic “information retrieval”.
The second view (Fig. 4) shows the publications
and co-authors that correspond to the research topic
“information retrieval.” The proposed topics are ob-
tained using the clustering engine Carrot (see sec-
tion 4) based on technical terms generated by the in-
formation extraction method described in section 3.
7 OUTLOOK AND FUTURE
WORK
In this contribution, we presented an approach to sup-
port information discovery tasks that combined tech-
nical term (TT) extraction, topic retrieval, and visu-
alization techniques. We introduced a new domain-
independent TT extraction method that allowed for
retrieving technical terms from paper abstracts with-
out using any additional domain specific information
(e.g., a lexicon or a seed-list). The extracted terms
are used for subsequent online-clustering of the query
results into topics. We illustrated a graph-based visu-
alization of an example co-author network that pro-
vided an opportunity for filtering the author’s publi-
cations and collaborators according to the topics ob-
tained through clustering of paper abstracts.
This example illustrates a clear advantage of the
combination of information extraction techniques and
interactive graph-based visualizations.
In the future, we plan to use the proposed TT ex-
traction method for detecting the retrieved TTs in the
body of a document. Then, we can concentrate only
on those passages that contain the found TTs. In do-
ing so, we can discover additional entities and rela-
tions that can be characteristic for a scientific paper
or a set of papers without processing the whole text
in an exceptionally efficient way. Such information
extraction techniques combined with interactive vi-
sualizations will enable a collaborative processing of
information by sharing it between a human and a ma-
chine.
ACKNOWLEDGEMENTS
The research project DILIA (Digital Library Assis-
tant) is co-funded by the European Regional Devel-
opment Fund (EFRE) under grant number 10140159.
We gratefully acknowledge this support.
REFERENCES
Anderson, T. D. (2006). Studying human judgments of rel-
evance: interactions in context. In Ruthven, I., editor,
Proceedings of the 1st international conference on In-
formation interaction in context, pages 6–14. ACM.
Barry, C. L. (1994). User-defined relevance criteria: An
exploratory study. Journal of the American Society
for Information Science, 45(3):149–159.
Bates, M. (1989). The design of browsing and berrypick-
ing techniques for the online search interface. Online
Review, 13(5):407–424.
Elmqvist, N. and Tsigas, P. (2007). Citewiz: A tool for
the visualization of scientific citation networks. Infor-
mation Visualization, 6(3):215–232. Technical Report
2004, published 2007.
Herman, I., Melancon, G., and Marshall, M. (2000). Graph
visualization and navigation in information visualiza-
tion: A survey. Visualization and Computer Graphics,
IEEE Transactions on, 6(1):24–43.
Mizukoshi, D., Hori, Y., and Gotho, T. (2006). Exten-
sion models of cone tree visualizations to large scale
knowledge base with semantic relations.
Robertson, G. G., Mackinlay, J. D., and Card, S. K. (1991).
Cone trees: animated 3d visualizations of hierarchical
information. In CHI ’91: Proceedings of the SIGCHI
conference on Human factors in computing systems,
pages 189–194, New York, NY, USA. ACM.
Spurk, C. (2006). Ein minimal berwachtes Verfahren zur
Erkennung generischer Eigennamen in freien Texten.
Diplomarbeit, Saarland University, Germany.
Vakkari, P. (1999). Task complexity, problem structure and
information actions - integrating studies on informa-
tion seeking and retrieval. Information Processing and
Management, 35(6):819–837.
KMIS 2009 - International Conference on Knowledge Management and Information Sharing
184

Yangarber, R., Winston, L., and Grishman, R. (2002). Un-
supervised learning of generalized names. In Proceed-
ings of the 19th International Conference on Compu-
tational Linguistics (COLING 2002).
Zhang, J., Mostafa, J., and Tripathy, H. (2002). Information
retrieval by semantic analysis and visualization of the
concept space of d-lib magazine. D-Lib
T M
Magazine,
8(10).
DILIA - A DIGITAL LIBRARY ASSISTANT - A New Approach to Information Discovery through Information
Extraction and Visualization
185
