
HIGH-LEVEL MODEL DEFINITION FOR MICROARRAY DATA
IN A FUTURE CLINICO-GENOMIC EHR
Anca Bucur, Jasper van Leeuwen, Richard Vdovjak and Jeroen Vrijnsen
Philips Research Europe, High Tech Campus 37,Eindhoven, The Netherlands
Keywords: Clinical information systems, Genomic-enabled EHR, Data models, Microarray, HL7, MIAME, MAGE.
Abstract: With the new discoveries in cancer research, increasing amounts of genomic data are starting to be used in
the context of the cancer patient management and should become part of the patient record. We propose a
high level data model for incorporating microarray data in a future genomic-enabled clinical information
system, based on existing and emerging standards. We argue that a genomic-enabled EHR system is
becoming highly relevant for clinical practice taking into account the new validated discoveries from
clinical research, but it could also have an important role to support new research by collecting and
integrating large amounts of data from clinical care and enabling extensive querying.
1 INTRODUCTION
The oncology clinical research and care are currently
faced with an explosion of relevant information. A
genetic disease, cancer requires increasing amounts
of genomic information for its diagnosis, patient
stratification and treatment selection. Much of this
currently expensive genomic data should become
persistent and accessible in a post-genomic clinical
information system, to be used for both patient
treatment and future research.
As more and more data of various types is
being collected for current clinical care, it becomes
increasingly important to preserve and ensure proper
access to that data to be used for research but also
for the future benefit of the patient, in the light of
new discoveries. There is a large body of genomic
information that will become part of standard care in
the coming years. As a medical record should
provide access to all relevant patient data used in the
process of delivering care, all the new genomic
information used in standard care, but also in
research, needs to make its way into the future
patient records. Preserving the data is especially
meaningful when storing and maintaining is
significantly cheaper than re-acquiring, and when
new insight can be obtained based on old data. Most
current off-the-shelf EHR solutions do not suitably
address the needs of clinical research (e.g. searching,
aggregation, integration) and they do not consider
the relevance of genomic data for clinical care.
In this paper we describe several scenarios
supporting the need for a genomic-enabled clinical
information system and propose an initial high-level
data model for microarray data in a future system.
As some of the standards currently have a research
focus and are very complex, in our model we
propose a simplified solution that is suitable to be
used in the context of a healthcare organization. This
work is part of the ACGT project (www.eu-acgt.org)
which aims to deliver to the cancer research
community an integrated clinico-genomic
environment enabling the sharing of data and of
biomedical research tools for clinical trials.
2 SCENARIOS SUPPORTING
THE NEED FOR PRESERVING
GENOMIC DATA
A typical hospital EHR does not provide a complete
view on all patient data and there are many legacy
systems still in use. A healthcare institution may
have for example an integrated system for the
inpatient environment, but not for outpatient. In
general, clinical researchers have their own
databases for research data. They share data on
collaboration basis, or when it is necessary for the
patient treatment.
Research data is maintained in different data
sources than the clinical data, where it can be
149
Bucur A., van Leeuwen J., Vdovjak R. and Vrijnsen J. (2010).
HIGH-LEVEL MODEL DEFINITION FOR MICROARRAY DATA IN A FUTURE CLINICO-GENOMIC EHR.
In Proceedings of the Third International Conference on Health Informatics, pages 149-154
DOI: 10.5220/0002592301490154
Copyright
c
SciTePress

properly managed and queried. This creates
unnecessary duplication and inconsistencies, as
patients treated in clinical trials will have data in
different systems.
With respect to genomic data, we have
identified several scenarios in which the
preservation of collected patient data is essential:
Genomic data is used for treating a concrete
patient, for assessment of prognosis or choice of
treatment.
A new discovery enables the use of existing
“old” data (accessible from the EHR) for treating
a concrete patient (for treating the same disease,
for treating other diseases, or for indentifying the
likelihood of relapse).
EHR-stored genomic data is used for research
when increased population is necessary (e.g.
research in rare diseases, detection of side effects
of drugs and treatment that are only visible in
large studies).
Existing “old” data is used for new hypothesis
building and testing.
Existing “old” data is used for revalidation
(refinement) of research results.
New techniques for analyzing the data are
applied (revalidation from another point of
view).
Exhaustive large-scale data mining of EHR data,
including genomic data (e.g. expression) to find
potentially relevant correlations.
In clinical practice, existing data is mined for
quality assurance to verify compliance with
guidelines, improving the process of delivery of
care and preventing errors.
In clinical research, existing data can be mined
for quality assurance to verify compliance with
trial protocols (e.g. mine trial data for quality
assuring matching, merge trials, retrospective
studies).
Genomic patient data is used to indicate potential
predisposition to disease (and the need for
testing) for other family members.
3 RELEVANT STANDARDS
In this section we discuss several standards relevant
in the context of modelling genomic data that should
become accessible from a post-genomic health
record system. These standards will be further used
in our model and in the description of the user
scenarios.
3.1 HL7 Reference Information Model
The main aim of the HL7 messaging standard is to
ensure that health information systems can
communicate their information in a form which will
be understood in exactly the same way by both
sender and receiver. Whereas HL7 version 2 was a
pure messaging standard for interoperability, version
3 (V3) not only specifies how to send a message, but
also what a message can contain. To achieve this
goal, V3 makes use of vocabularies and ontologies
like SNOMED (SNOMED, 2008) and LOINC
(LOINC, 2009). At the basis of all HL7 V3
messages is the Reference Information Model (RIM)
(HL7 RIM, 2009), an abstract model of the concepts
which underlie healthcare information.
3.2 HL7 Clinical Genomics
The Clinical Genomics working group develops
HL7 V3 standards that enable the exchange of
interrelated clinical and personalized genomic data
between interested parties (HL7 CG, 2008).
Currently, the domain consists of three topics:
Genotype, Genetic Variation, and Pedigree. The
latter topic aims at describing a patient’s heredity
based on genomic data. It utilizes the models from
the Genotype topic to contain the genomic data for
the patient’s relatives.
The Genotype topic consists of two (HL7 RIM-
based) models: the Genetic Locus and the Genetic
Loci. The latter model groups several genetic locus
instances, e.g. in case of a genetic test of several
genes. The first model describes data related to a
genetic locus: the position of a particular given
sequence in a genome or linkage map. The model
includes sequencing and expression data, and can be
linked to clinical information or phenotypes.
Existing bioinformatics mark-up languages such as
MAGE-ML (MAGE, 2006) and BSML (BSML,
2001) are used to represent the raw genomic data.
The Genetic Variation topic defines a model that
is a constraint over the Genotype topic models. The
focus of this model is on variations in the DNA of
individuals, derived using methods such as SNP
probes, sequencing and genotype arrays that focus
on small scale genetic changes. However, gene
expression analysis, e.g. based on microarray data, is
not suitable for the Genetic Variation model and will
be addressed (in the near future) by different models
within the HL7 Clinical Genomics working group.
HEALTHINF 2010 - International Conference on Health Informatics
150

3.3 Minimum Information about a
Microarray Experiment (MIAME)
Although increasingly used for gene expression data
analysis at a genome-wide level, microarray
technology still has the limitation of insufficient
standardization for presentation and exchange of
such data. The MIAME standard (Brazma, 2006)
aims at establishing a common way for recording
and reporting microarray-based gene expression
data, and proposes the minimum information
required to ensure that microarray data can be
interpreted and that the results that yield from the
analysis of the data can be independently verified.
The standard only defines the content and the
structure of the information and does not address the
actual technical format of storing and
communicating the data.
MIAME has also identified the need for
controlled vocabularies and ontologies for data
representation in order to enable interoperability. As
there is a very limited availability of suitable
controlled vocabularies, MIAME proposes a
representation in lists of ‘qualifier, value, source’
triplets, which authors can use to define their own
attributes (i.e. qualifiers) and provide the appropriate
values and the source from which the terms were
extracted.
A significant amount of context data is necessary
to describe a microarray experiment because the
results of such an experiment (gene expression) are
only meaningful in the context of the conditions in
which the experiment was run. Most microarray
experiments only report relative changes in gene
expression relative to a non-standardized reference,
the data is normalized in different ways and is
represented in non-standardized formats, and the
annotation describing the data is often insufficient.
All these factors make comparing data from distinct
experiments very difficult. MIAME attempts to
alleviate these issues by specifying the annotation
necessary to properly interpret the data and the
detailed description of the experiment, including the
way in which the gene expression level
measurements were obtained.
Next to the gene expression matrix, which
contains for each gene and sample in the array the
measured expression, MIAME advises to provide
information about the genes whose expression was
measured and about the experimental conditions
under which the samples were taken. The
information required can be divided at a conceptual
level into three logical parts: gene annotation,
sample annotation and a gene expression matrix.
3.4 MAGE
While MIAME focuses on the conceptual content of
the data, specifying what information is needed in
order to be able to interpret and reproduce a
microarray experiment, MAGE (MAGE, 2006)
delivers data exchange standards to facilitate the
exchange of gene expression data. The core of
MAGE is MAGE-OM, which provides an object
model for the exchange of gene expression data.
MAGE also proposes two data exchange formats,
MAGE-ML – which provides a mark-up language-
and MAGE-TAB – which provides a tabular format
(which is the current recommendation). MAGE-OM
(OMG, 2003) defines the object model for gene
expression data and it is modelled using UML.
The model can express microarray designs,
microarray manufacturing information, microarray
experiment setup and execution information, gene
expression data, and data analysis results, satisfying
the MIAMI requirements. MAGE-OM tries to be
generic and as complete as possible. Users typically
use a subset of the provided classes and relations,
which would fulfil their needs.
MAGE-ML captures MAGE-OM in an xml
notation, explicit mapping rules map the MAGE-
OM model to xml. Although MAGE-ML is
supported in various tools as import and export
format, it is a cumbersome format to use in a
laboratory when no appropriate tooling or software
development expertise is available.
MAGE-TAB (Rayner, 2006) fills this gap by
providing a simple format, still capturing the
requirements of the MIAMI standard. MAGE-TAB
is a tabular format and can be easily manipulated
with various tools (even spreadsheet programs).
4 HIGH-LEVEL DATA MODEL
FOR MICROARRAY DATA
Although there are several existing HL7 standards
addressing the issues of communication of clinico-
genomic data, we consider them not applicable for
the actual storage of the data. On the other hand, the
MIAME and MAGE standards provide models for
the storage and exchange of microarray-based gene
expression data, but are mainly tailored towards
research purposes: The underlying data models are
too complex and too elaborate to be directly used in
an EHR. For that reason, we define an initial
simplified model for the storage of genomic
information in a medical record, which combines
HIGH-LEVEL MODEL DEFINITION FOR MICROARRAY DATA IN A FUTURE CLINICO-GENOMIC EHR
151
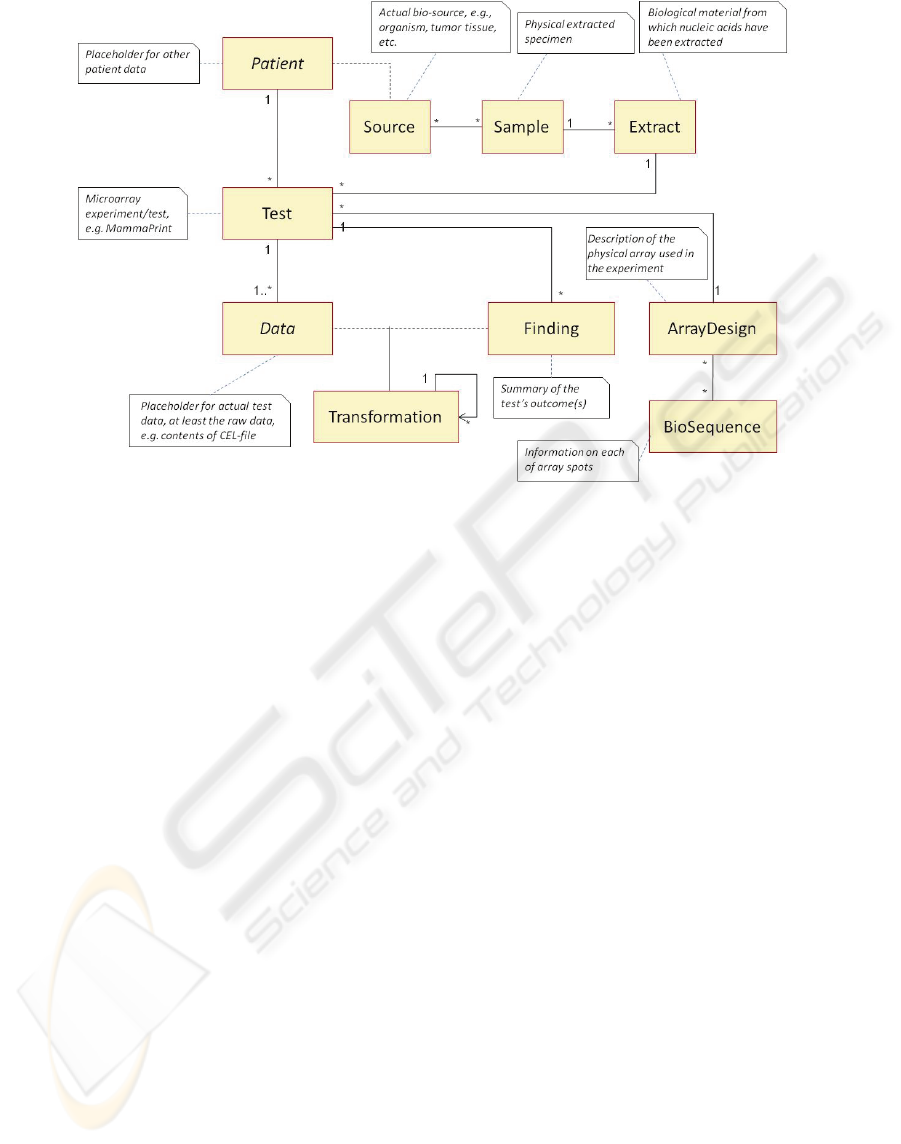
Figure 1: High-level data model described as an UML class diagram.
elements from existing standards with the
requirements captured in our scenarios. The aim of
this model is to provide a higher abstraction to
genomic data and microarray data in particular such
that it shields the user from the underlying
complexity of the involved standards, while clearly
identifying their use and application. The resulting
high-level model is depicted in Figure 1.
In this model, we assume a patient-centric
medical record, where all clinical data as well as the
demographic data is encapsulated by the Patient
class. The scope of the high-level model is limited to
microarray data; all other relevant medical as well as
administrative data, such as the purpose of ordering
a microarray test or an overview of the patient’s
medical history, is reachable from the abstract
Patient class. Similar to other clinical tests, a doctor
can order multiple microarray-based tests for a
patient. The Test class stores all metadata on the
performed test, such as the type and the purpose of
the test, the date of testing, and other experimental
factors.
The actual microarray used for the test is
described in the ArrayDesign class. As more and
more standardized microarray tests become available
in the future, ArrayDesign can simply be an external
reference to the design as published by the
manufacturer, or it can be the complete array’s
blueprint, including information on each of the
sequences (BioSequence class) placed on the spots
of the array.
Because the microarray test only has a meaning
in the context of a particular extract that was
hybridized onto the array, we describe this extract,
i.e., the biological material for which the genetic
profile is determined, in the Extract class. In this
class, it is possible to describe how the extract was
derived from the physical extracted specimen, the
sample (Sample class). The source from which the
sample originates (e.g., from the lung, left breast,
etc.) is described in the Source class.
The outcome of the test is described in the
Finding class. This can be a single finding, for
example whether there is a good or bad prognosis,
but also multiple findings are possible, depending on
the type of array used. For example, the
progesterone, oestrogen and HER2 expression levels
could be measured along the lines of the research
described in (Roepman, 2008), generating several
findings in a single test. In this initial model we
define the findings simply as key-value pairs, as
each finding is test-specific and no standardization
of microarray test results exists yet.
Each of the clinical findings is derived from a
particular dataset of the test, by applying a specific
(algorithmic) transformation(s) on that data. This
can be a simple transformation, e.g., a threshold
function, or a complex transformation consisting of
a sequence of smaller transformations. The
Transformation class is used to encapsulate each of
these data processing steps.
For research purposes, it is necessary to also
HEALTHINF 2010 - International Conference on Health Informatics
152

Data
ArrayDesign
Transformation
Figure 2: Mapping from MAGE BioAssayData package to our data model.
store the actual (raw) data of the test, possibly with
some additional description or annotation. The Data
class is specified here as an abstract class, as there
are multiple types of data that can be stored.
Examples of the data to store are the intensity values
and the scanned images. The MAGE standard
defines how to store the data in its BioAssayData
package. As this package actually defines more than
what we intend with the abstract Data class, Figure 2
shows the mapping from this package to our model.
5 MODEL WALKTHROUGH
In Section 4 we have identified several clinically
relevant scenarios which pose their own
requirements on the data model. In this section we
revisit one of these scenarios: New discovery
enables the use of existing “old” data for treating a
patient, and map it onto our initial data model,
identifying the relevant classes and their role. The
classes that are used are highlighted by rounded
rectangles. In this scenario, a new discovery is made
which refers to the same genomic data that has been
collected for a previously ordered test.
In Figure 3 we adopt the following color
convention: Dark color indicates previously created
class instances (e.g. previously stored data); light
color indicates newly created instances (e.g. a new
test or transformation).
The relevant patient clinical data is accessed via
the Patient class. Classes Source, Sample and
Extract are only accessed to verify the metadata
about the physical entities they represent – if the
metadata conforms to the newly designed test, there
is no need to redo the sample extraction/biopsy.
The new test represented by the Test class can
access the previously stored data via the Data class.
The new test introduces a sequence of new
transformations represented by the Transformation
class, which ultimately produces a new finding
captured by the Finding class. In order to be able to
understand and process the stored data, in some
cases the microarray design of the previous test
needs to be consulted.
HIGH-LEVEL MODEL DEFINITION FOR MICROARRAY DATA IN A FUTURE CLINICO-GENOMIC EHR
153

Figure 3: Model walkthrough.
6 CONCLUSIONS
With new discoveries in cancer research, increasing
amounts of genomic data are starting to be used in
the context of the cancer patient management and
should become part of the patient record. For clinical
research the data collected in the clinical practice is
a valuable resource allowing for new hypotheses
generation and testing, research in rare diseases,
selection of patients for trials, etc. In this context,
storing all genomic information available and not
only the processed test results, even when that
information is not immediately used in the care-
providing process, is meaningful.
As the costs of generating genomic data, such as
microarray-based gene expression, and extracting
information from that data is still quite high it does
not make economic sense to discard all the collected
data and to preserve only the test result instead of
storing and re-using it, especially that it is
increasingly clear that genomic data can generate
much more knowledge than what can be extracted in
a single test or experiment and that the
understanding of the expression data evolves.
Additionally, it may also be the case that it is not
possible to re-run a relevant test (e.g., at recurrence
of disease data from previous occurrence may be
relevant).
New standards for storing and exchanging
genomic data such as MIAME and MAGE also
require that all data should be preserved and
annotated, including the raw microarray image. Of
course, MIAME’s and MAGE’s main focus is
research, but the reasoning behind the storage and
full annotation of all genomic data is precisely based
on the fact that the data encapsulates information far
beyond the scope of a single experiment and should
be preserved and shared.
In this paper we have proposed a high-level data
model for microarray data in a future clinical
information system, based on established standards.
As the available standards for microarray data
currently have a research focus and are very
complex, in our model we propose a simplified
solution that is suitable to be used in the context of a
healthcare organization.
REFERENCES
HL7 Reference Information Model, 2009. Available at
http://www.hl7.org/v3ballot/html/infrastructure/rim/ri
m.htm, last viewed 2009.
SNOMED CT, 2008. Available from
http://www.ihtsdo.org/snomed-ct/release-of-snomed-
ct/.
LOINC, 2009. Available from http://loinc.org/.
HL7 Clinical Genomics Domain, 2008. Available from
http://www.hl7.org/v3ballot/html/domains/uvcg/uvcg.h
tm, last viewed 2009.
MicroArray Gene Expression, 2006. Available from
http://www.mged.org/Workgroups/MAGE/introduction
.html, last viewed 2009
Bioinformatics Sequence Markup Language, 2001.
Available from http://xml.coverpages.org/bsml.html,
last viewed 2009.
Brazma, A. et al., 2001. Minimum information about a
microarray experiment (MIAME) – toward standards
for microarray data. In Nature Genetics, vol. 29, 365-
371, 2001.
Gene Expression Specification, Object Management
Group, 2003. Available from http://www.omg.org/cgi-
bin/doc?formal/03-02-03, last viewed 2009
Rayner, T.F. et al., 2006. A simple spreadsheet-based,
MIAME-supportive format for microarray data:
MAGE-TAB. In BMC Bioinformatics, vol. 7, 2006.
Roepman, P. et al., 2008. Microarray-based readout of ER,
PR, and HER2 expression in breast cancer tissue. In
Breast Cancer Symposium, 2008.
HEALTHINF 2010 - International Conference on Health Informatics
154
