
AN INFORMATION FILTERING SYSTEM FOR E-HEALTH
The Health-on-Net Experience
Nicola Capuano, Matteo Gaeta, Vincenza Precone
Dept. of Information Engineering and Applied Mathematics, University of Salerno, Italy
Mario Scherillo
NEXERA S.c.p.A, Napoli, Italy
Keywords: Text Categorization, ICD, User Profiling, Information Filtering, Matchmaking.
Abstract: This paper describes a work performed in the framework of the HealthOnNet project purposed to define and
implement an Internet-based repository of diagnostic exams and medical reports connecting several Italian
hospitals. The repository, which will be used as an historical and legal archive of clinical data, offers second
opinion teleconsulting features as well as advanced categorization and filtering services. The paper is
focused on this latter point and describes the process and the algorithms we defined to automatically classify
medical documents (with respect to the widely adopted International Classification of Diseases and Related
Health Problems of the World Health Organization) and to filter them on the basis of a user defined profile.
Then it describes the developed prototype and some experimentation results.
1 INTRODUCTION
Healthcare is evolving in a specialised environment
where the patient doesn’t select the nearest hospital
but the best one suited to the suffered disease. For
this reason he will write several pieces of his health
curriculum on several different “books” that will so
often results fragmented, misaligned and discordant.
In this context, the HealthOnNet (HoN) project’s
purpose was to build an Internet-based repository
connecting several Italian hospitals and able to store
and index medical documents of several kinds like
diagnostic exams and medical reports. This will act
as a centralized dossier collecting and connecting
every patient’ clinical experience.
The HoN repository represents an historical and
legal archive of clinical data accessible through a
Web browser by medical doctors as well as directly
by patients. It is also able to foster cooperation
among hospitals by allowing easy exchange of data
and enabling second opinion teleconsulting.
Moreover the repository can also be exploited by
researchers to collect information about pathologies
and their territorial distribution as well as about
applied medical treatments and their outcomes. This
can be useful e.g. for evidence based medicine
(Elstein, A.S., 2004) whose aim is to suggest health
care practices based on a wide set of reliable data
about clinical outcomes.
In order to support this latter feature we defined
a process and a set of algorithms for the automatic
categorization of medical documents with respect to
the widely adopted International Classification of
Diseases and Related Health Problems (ICD) of the
World Health Organization (WHO).
Moreover we defined profiling techniques to let
researchers define their interests in terms of ICD
categories and developed a system able to retrieve
relevant documents on the repository according to
the defined profile. Finally we also defined and
developed matchmaking algorithms to find and put
in touch researchers with similar interests.
This paper describes achievements made about
these topics within HoN. Section 2 presents related
literature while sections 3 and 4 deepen theoretical
issues and describe defined text categorization, user
profiling and matchmaking algorithms. Section 5
presents the developed prototype and shows some
preliminary experimentation results. Finally section
6 adds some brief concluding remarks.
210
Capuano N., Gaeta M., Precone V. and Scherillo M. (2010).
AN INFORMATION FILTERING SYSTEM FOR E-HEALTH - The Health-on-Net Experience.
In Proceedings of the Third International Conference on Health Informatics, pages 210-215
DOI: 10.5220/0002697402100215
Copyright
c
SciTePress

2 RELATED WORK
Internet and local networks have greatly increased
the availability of medical information and reduced
the cost and the time needed to access it. Examples
of on-line information related to the health domain
include medical reports, bibliographies, conference
proceedings, clinic instructions, health organisation
information, discussion forums, etc.
This represents not only an opportunity but also
a problem for users with respect to the huge volume
of information that is generated daily. In order to
find relevant information in this context the adoption
of controlled vocabularies for information indexing
as well as of advanced categorization and filtering
techniques is more and more needed.
With respect to the first topic, several controlled
vocabularies for medicine exist. As an example, the
World Health Organization (WHO) defined the
International Classification of Diseases and Related
Health Problems (ICD) that provides codes to
classify diseases and a variety of signs, symptoms,
abnormal findings, complaints, social circumstances
and external causes of injury or disease. The current
version of ICD (WHO, 2004) is composed of more
than 155.000 codes grouped in 21 main chapters.
Moreover, the U.S. National Library of Medicine
developed the Medical Subject Headings (MEsH), a
controlled vocabulary of terms aimed at indexing
papers and books in life sciences and serving as a
thesaurus to facilitate searches in the medical field
(NLM, 2008).
MeSH includes more than 100.000 concepts and
a hierarchy with more than 11 deepening levels. It is
used to index MEDLINE, the biggest database of
medical information on-line. Several systems have
been developed to find in it relevant information like
the Grateful Med System, a windows-based querying
user interface and COACH, a system that converts
keywords and phrases in MeSH compatible queries.
Unfortunately these systems require a learning
phase that still discourages potential users. Moreover
they are not able to actively support users during the
search task by trying to determine user interests and
to refine searches by considering this information. In
other word they are not filtering systems.
An information filtering system is able to select
from an information source only those items that are
relevant for a given user basing on a (explicitly
defined or implicitly inferred) user profile. A user
profile describes the interests of a user with respect
to domain topics and may be generated by exploiting
machine learning techniques e.g. by extracting key-
words from a set of relevant documents and/or by
collecting user (positive or negative) feedback about
documents suggested by the system.
One of the few existing filtering systems in the
medical domain is Kavanah (Santos, E., Nguyen,
H., 2000) that is based on interface agents that learn
the interests and the preferences of the users while
they perform search tasks. Discovered interests and
preferences are then exploited to help users retrieve
further information and relevant knowledge.
The system we propose in this paper tries to
merge the advantages coming from the application
of a controlled vocabulary (ICD) together with those
coming from a filtering system. It is able, from one
side, to automatically categorize unknown medical
documents on the basis of ICD and, from the other
side, to filter relevant information on the basis of an
explicit ICD-based user profile.
For these reasons it is different from existing
search tools based on controlled vocabularies (like
Grateful Med System and COACH) because it also
offers profile-driven searches. It is also different
from traditional filtering systems (like Kanavah)
because available documents are pre-classified with
respect to a controlled vocabulary. This not only
empowers search facilities but also ensures a greater
interoperability with external systems based on such
vocabularies.
3 THE CATEGORIZATION
METHODOLOGY
The categorization process we apply is based on
machine learning techniques enabling the automatic
building of a classifier by letting it learn distinctive
features of the categories of a given domain starting
from a training set of pre-classified documents
(Sebastiani, F., 2002).
In other words, given a certain set of categories
(those provide by ICD in our case), a training set is
used to extract the distinctive features (or terms) of
each category. At this point, once a new document is
presented to the system, it will be able to assign it to
none, one or more categories on the basis of the
value of a given similarity function.
More formally, given a document domain D and
a category domain C, the purpose of the algorithm is
to build a function
φ
able to associate each pair (d, c)
with d ∈ D and c ∈ C a membership value of d in c
included in the range [0, 1].
As anticipated, our approach relies on an initial
corpus Ω ⊂ D of pre-classified documents. In such
sense every pair (d, c) with d ∈
Ω
so that
φ
(d, c) = 1
is a positive sample for the class c while every pair
(d, c) so that
φ
(d, c) = 0 is a negative sample for the
AN INFORMATION FILTERING SYSTEM FOR E-HEALTH - The Health-on-Net Experience
211
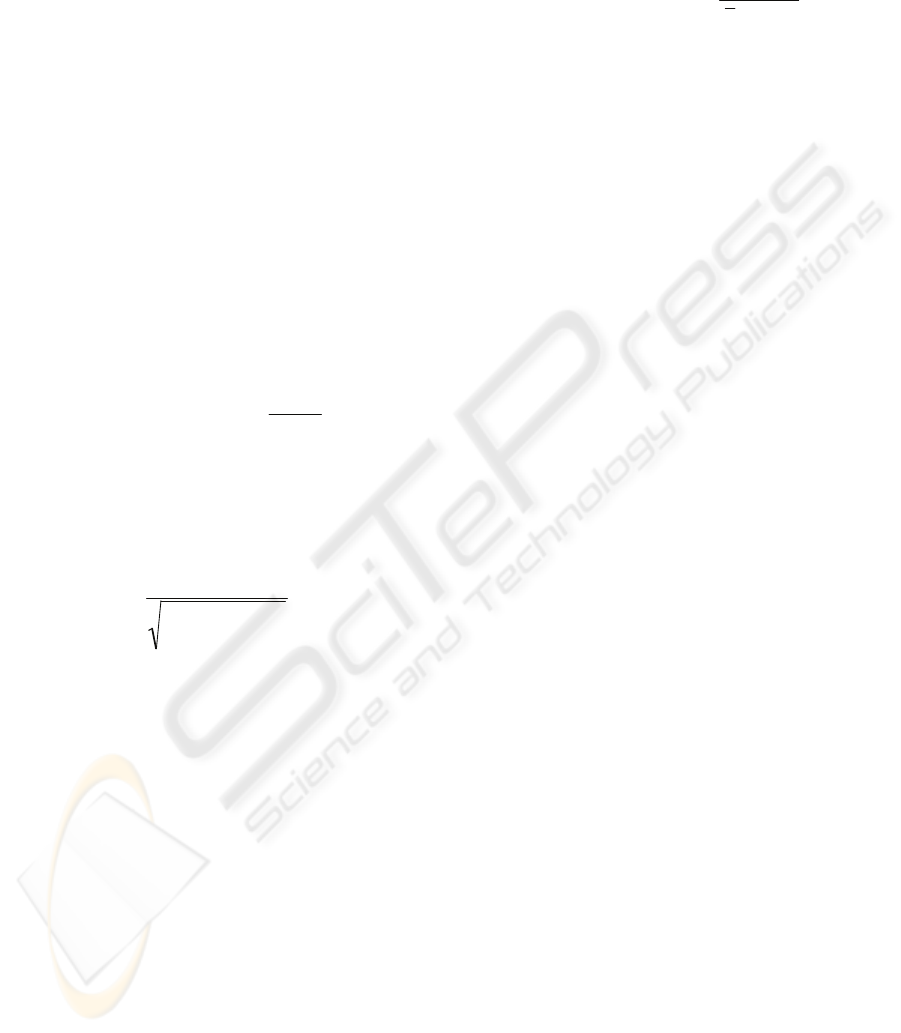
same class c.
To classify unknown documents (so belonging to
D – Ω) it is necessary to identify relevant terms from
each known document and, by considering their
membership to the classes of C, to understand the
relevant terms representing each class. In such a way
the classification of an unknown document can be
done by extracting relevant terms from it and by
associating it to the class represented by the most
similar terms.
To extract relevant terms from a given document
d means to transform it in an array of weights dw so
that dw = (w
1
, …, w
|T|
) where T is the set of terms
appearing at least once in a document of Ω and each
w
i
(so that 0 ≤ w
i
≤ 1) represents how much the term
t
i
belonging to T represents the semantics of d.
In order to calculate the weight of the i-th term t
i
in a given document d, we chose to use the standard
tf-idf (term frequency inverse document frequency)
function as defined in (Salton, G. and Buckley, C.,
1988):
tfidf (t
i
,d) =#(t
i
,d) ⋅ log
| Ω |
#
Ω
(t
i
)
(1)
where #(t
i
, d) indicates how many times the term t
i
appears in the document d while #
Ω
(t
i
) indicates the
number of documents in Ω where t
i
appears at least
once. In order to normalise weights in the range
[0,1] we chose to define each weight w
i
as follows:
w
i
=
tfidf (t
i
,d)
(tfidf (t,d))
2
t ∈T
∑
. (2)
To speed up the calculation of the arrays of
weights and to improve the classifier performances,
we apply the following pre-processing algorithms:
tokenization to transform a document in a list of
words (removing figures, symbols, tags, etc.), stop
words removal (to remove not relevant terms like
articles, conjunctions, pronouns, prepositions) and
stemming (to reduce an inflected or derived word to
its stem, base or root form) (Van Rijsbergen, C.J. et
al., 1980).
Once all arrays of weights associated to training
documents are available, it is necessary to calculate
the degree of membership of each new document to
one of the selected categories. To do that we use a
Bayesian classifier (Lewis, D.D., 1998) that is
performing also with small training sets (Sebastiani,
F., 2002).
This kind of classifier sees the function
φ
(d, c)
in terms of conditional probability that a document d
belongs to the category c given an associated array
of weights dw = (w
1
, …, w
|T|
). Assuming that all the
weights are statistically independent, it is possible to
estimate the function
φ
(d, c) as follows:
log(
φ
(d,c)) = w
i
log
p
i
(1 − p'
i
)
p
i
(1 − p
i
)
i=1
|T |
∑
(3)
where w
i
is the i-th weight related to the document d,
p
i
= P(w
i
= 1 | c) is the conditional probability that a
document contains the term t
i
given that it belongs to
c and p
i
’ = P (w
i
= 1 | c’) is the conditional proba-
bility that a document contains the term t
i
given that
it belongs to a category different from c.
By applying (3), in order to define a classifier for
each category c, it does suffice to estimate p
i
and p
i
’
for each i so that 1 ≤ i ≤ |T| starting from training set.
We give more details about that in next paragraphs.
3.1 The Training Phase
This phase is based on the defined methodology and
includes the so called “three characters” categories
of ICD (WHO, 2004) in the category set C. The
training set Ω is instead composed by 5372 pre-
classified medical documents. We have chosen ICD
given its widespread adoption and given that a lot of
pre-classified documents are already available on-
line and can be used to train the classifier.
The training phase works as follows: for each
document d of the training set, the tokenization step
generates a list of terms td from which the algorithm
removes terms belonging to a stop-list. Remaining
terms are stemmed to obtain their base form.
Then the algorithm generates the set T with all
terms occurring at least once in td for more than 3
documents of the training set (so we set
ρ
= 3). For
each document the array dw = (w
1
, …, w
|T|
) is then
calculated by applying (1) and (2).
After that, for each category c belonging to C,
the algorithm calculates the parameters p
1
, …, p
|T|
where each p
i
is the ratio between the number of c
members containing t
i
and the number of c members.
Then it calculates the parameters p
1
’, …, p
|T|
’ where
each p
i
’ is the ratio between the number of members
of all classes different from c containing t
i
and the
number of members of all classes different from c.
After that the classifier is trained and ready to be
used as we describe in the following paragraph.
3.2 The Classification Phase
After the training phase, it is possible to classify any
document of the HoN repository by applying the
following steps.
HEALTHINF 2010 - International Conference on Health Informatics
212
For each available document d in the repository,
relevant terms are extracted (through tokenization,
stop words removal and stemming) and the array of
weights dw = (w
1
, …, w
|T|
) is calculated applying (1)
and (2). Then the membership function
φ
(d, c) is
calculated for each category using (3). Classification
results are stored and the process is repeated for any
new document that becomes available.
Once the
φ
(d, c) is estimated for any document
of the repository, users may chose to see categories
related to each document (ordered downward on
φ
)
or documents belonging to each category (ordered
downward on
φ
too). In both cases, only categories
or documents with
φ
greater then a given threshold
are considered.
4 MATCHMAKING AND
PROFILING
The developed prototype applies explicit profiling in
order to let each user specify ICD categories it is
interested in. As already said all “three characters”
categories are supported as well as the 21 main
chapters (each grouping several categories).
A user can define his or her profile by selecting
one or more categories and/or one or more chapters.
If a chapter is selected then all categories belonging
to it are implicitly selected.
Once the profile is defined, the system suggests
to the user all the documents from the repository that
belong to categories of interest ordered according to
the function
φ
. Once a new document is added to the
repository it is classified and, if relevant with respect
to a user profile, suggested.
Starting from defined profiles, the system is also
able to apply a matchmaking algorithm to identify
and put in touch users with similar interests (so with
similar profiles). Given the category domain C, a
profile p can be defined as a function p: C → {0, 1}
where p(c
i
) = 1 if the user selects the category c
i
and
p(c
i
) = 0 otherwise.
In order to take into account chapters, we define
C’ as the superset of C including all categories and
chapters and define the “modified” profile function
p’: C’ → {0, 1} so that p’(c
i
) = p(c
i
), if c
i
∈ C and
p’(c
i
) = mean { p(c
j
) | c
j
belongs to c
i
} otherwise.
Given two users with two profile functions p and
q we calculate their similarity s(p, q) in this way:
s(p,q) =1−
′
p (c
i
) −
′
q (c
i
)
i=1
n
∑
max
′
p (c
i
),
′
q (c
i
)
()
i
=
1
n
∑
. (4)
When the similarity degree between two users is
greater then a give threshold, if both users agree,
they are automatically put in touch by the system.
5 PROTOTYPE DEVELOPMENT
AND EXPERIMENTATION
On the basis of the theoretical results described we
developed and experimented a filtering prototype for
medical documents in the HoN repository.
In the following paragraphs we provide details
about achievements obtained in these three phases.
5.1 Prototype Architecture
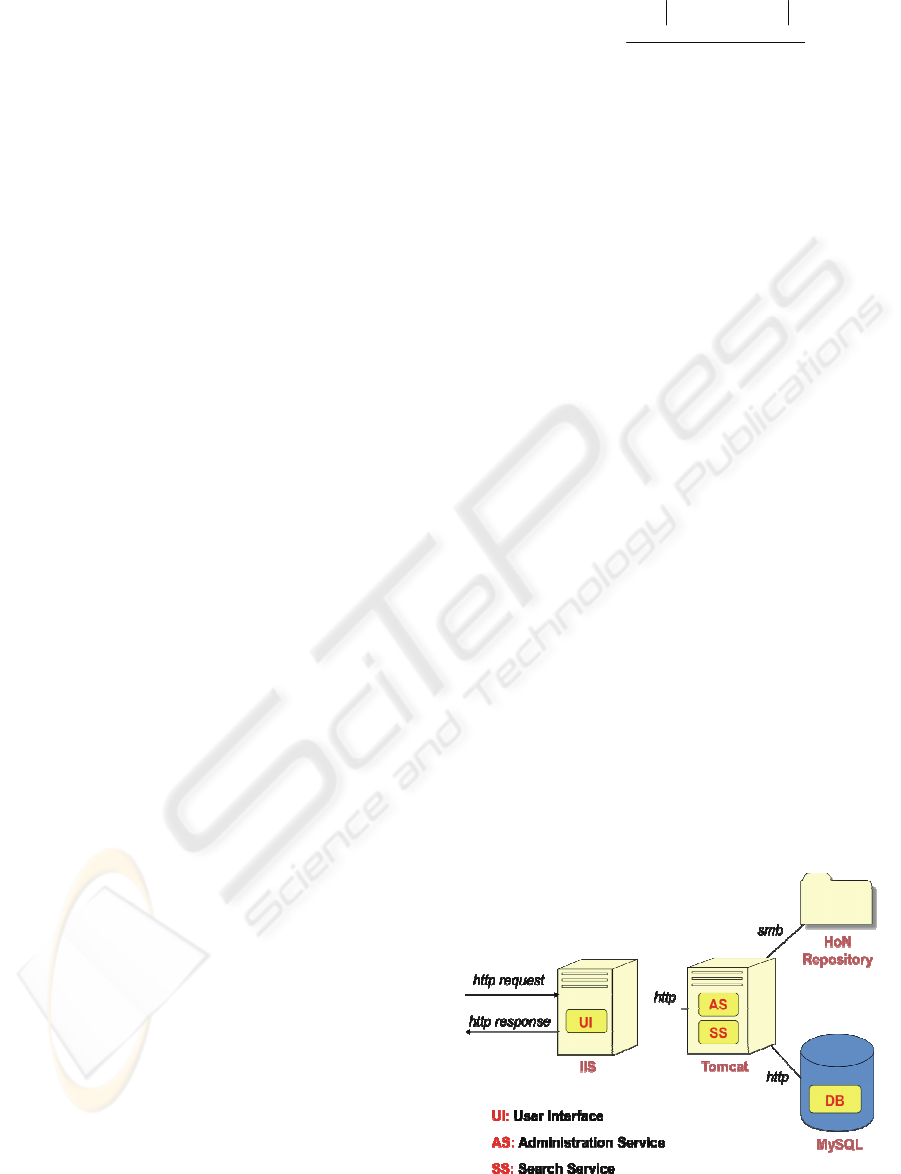
As depicted in figure 1, the developed prototype is
made of three components: a Web application and
two Web services.
The User Interface (UI) is a Web application
allowing medical doctors and administrators
to access system functionalities.
The Administration Service (AS) is a Web
service providing functions to manage training
and categorization phases as well as document
indexing and system configuration.
The Search Service (SS) is a Web service
providing functions for document retrieval
(with respect to a given user profile or to a full
text search) and for matchmaking.
System functions are accessed by users through
UI. Basic functions are directly handled by UI but
Figure 1: The prototype architecture (deployment view).
AN INFORMATION FILTERING SYSTEM FOR E-HEALTH - The Health-on-Net Experience
213
for more complex functions, UI simply forwards
requests to AS and/or SS, then it composes and
displays the obtained results.
Available documents are captured directly on the
HoN repository. A synchronization function, able to
find and classify new documents and remove records
for deleted documents is periodically invoked.
5.2 Prototype Usage
The first step needed to use the prototype is the
authentication through the UI. Each user has a role
(administrator or doctor) so, after the authentication,
he is redirect to the administrator or to the doctor
home page, depending on the role.
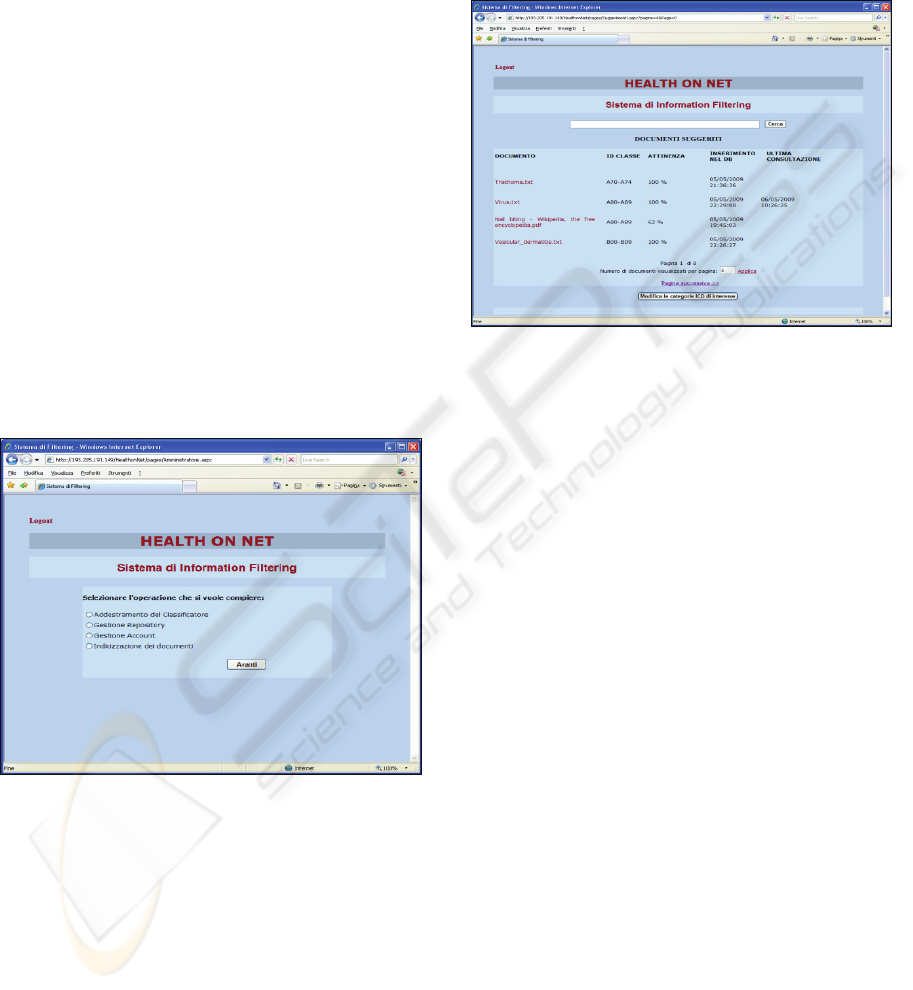
Figure 2 shows the administration home page
whose accessible functionalities are:
system training (to configure and to launch the
training phase of the categorization process);
document repository management (to select and
synchronize the document repository);
account management (to add, delete or modify
system users and to assign roles);
document indexing (to enable full-text search
by indexing repository documents).
Figure 2: The administration home page.
Figure 3 shows the medical doctor home page
whose accessible functionalities are described in the
following list.
definition of categories of interest (they must be
defined the first time the doctor accesses to
the system and can be modified anytime);
accessing to the list of suggested documents
(ordered downward with respect to calculated
relevance with respect to the profile);
searching for new documents (a full featured
keyword based search engine is also provided
to complement filtering functions);
opening and reading a selected or suggested
document;
matchmaking (i.e. accessing the list of system
users whose profile is similar to the one of the
current user).
Figure 3: The medical doctor home page.
When a doctor accesses the system for the first
time, he must select his categories of interest. Once
he has done that, the system is able to show in his
home page a list of documents related to selected
categories. Each time a new document is added to
the HoN repository, it is automatically suggested to
interested users. Accesses to available documents
are registered and shown in the document list.
In order to take into account not only long-term
interests but also short-term needs, the system also
allows to make full text queries. The query manager
supports Boolean (+ and -) as well as logical (AND,
OR, NOT) operators as in common search engines.
5.3 Prototype Experimentation
To experiment the filtering system we have built a
classifier for the 226 “three characters” categories of
ICD and we have trained it with 5372 pre-classified
documents coming from Wikipedia.
We have used documents taken from Wikipedia
(that came pre-classified with respect to ICD-10) to
avoid, at least in a first stage, the involvement of any
domain expert. By using a simple spider following
medical links on Wikipedia we were able to obtain
more then 10 training documents for each category.
Once the classifier was ready, we performed the
synchronization phase to classify a subset of medical
documents included in the HoN repository. Then we
involved a medical researcher that defined a profile
and started to browse system suggested documents
HEALTHINF 2010 - International Conference on Health Informatics
214

providing positive feedback on the system ability to
retrieve relevant documents and on the friendliness
of this first version of the user interface.
The next step is to involve experts in the training
phase in order to build a significant document set to
initialize the classifier. Then a formal evaluation of
system performances will be made by calculating
classical indicators like precision (i.e. the number of
relevant documents retrieved divided by the number
of documents retrieved) and recall (i.e. the number
of relevant documents retrieved divided by the total
number of existing relevant documents).
6 CONCLUSIONS AND FUTURE
WORK
The purpose of this paper was to describe a filtering
tool for medical information built upon a repository
of diagnostic exams and medical reports collecting
documents from several Italian hospitals. Filtering
was aimed to support researchers in the collection of
relevant data on pathologies distribution, applied
medical treatments and their outcomes.
The developed filtering prototype can categorize
unknown medical documents on the basis of the ICD
classification and can filter relevant documents on
the basis of ICD-based user profiles (nevertheless it
can be configured to handle other classifications). It
so merges advantages coming from the adoption of a
controlled vocabulary (vocabulary-based searches,
tools interoperability, etc.) with advantages coming
from the application of an information filtering
approach (e.g. profile-based searches).
The defined categorization process is composed of
two phases: a training phase aimed at building a
classifier through the adoption of machine learning
techniques basing on the analysis of a set of training
documents and a classification phase where the so
built classifier is used to classify new documents.
Moreover we defined profiling techniques to let
researchers explicitly define their interests as well as
matchmaking algorithms to find and put in touch
researchers with similar interests. Defined process
and related algorithms have been implemented and
integrated within the HoN repository. Preliminary
experimentation results are satisfactory.
Future work about these topics will be focused on
adding implicit profiling techniques based on the
observation of user behaviour as well as the use of
relevance feedback to improve, from one side, the
adherence of user profiles to real user interests and,
from the other side, performances of the document
categorizer.
In this way the system will be able to consider
not only current interests of each user but also their
evolution over time without the need of redefining
them by hand. A large scale experimentation is also
foreseen to evaluate system advantages at a greater
extent, also with respect to similar systems.
ACKNOWLEDGEMENTS
This work was partially founded by the Campania
Region under the HealthOnNet project conducted by
the Dept. of Information Engineering and Applied
Mathematics of the University of Salerno together
with the society Nexera S.c.p.a.
REFERENCES
Elstein, A.S., 2004. On the origins and development of
evidence-based medicine and medical decision making
In Inflammation Research, vol. 53 suppl. 2. Springer.
Lewis, D.D., 1998. Naive (Bayes) at forty: The indepen-
dence assumption in information retrieval. In Proc. of
the 10
th
European Conference on Machine Learning
(ECML-98), Chemnitz, Germany, pp. 4-15.
NLM, 2008. Introduction to MeSH, U.S. National Library
of Medicine, http://www.nlm.nih.gov/mesh/.
Pratt, A., Sim, I., 1995. Physician’s Information Customi-
zer (PIC): Using a Shareable User Model to Filter the
Medical Literature. In Proc. of the Int’l Conf. on
Medical Informatics MEDINFO’95, pp.1447-1451.
Salton, G., Buckley, C., 1988. Term-weighting approaches
in automatic text retrieval in Information Processing
& Management, vol. 24, no. 5, pp. 513–523. Elsevier.
Santos, E., Nguyen, H., 2000. Medical Document Infor-
mation Retrieval through Active User Interfaces. In
Proceedings of the 2000 International Conference on
Artificial Intelligence (IC-AI '2000), Las Vegas, NV.
Sebastiani, F., 2002. Machine Learning in automated Text
Categorization. In ACM Computing Surveys, vol. 34,
no. 1, pp. 1–47. ACM .
Van Rijsbergen, C.J., Robertson, S.E., Porter, M.F., 1980.
New models in probabilistic information retrieval.
British Library. London.
WHO, 2004. The International Statistical Classification of
Diseases and Related Health Problems ICD-10, World
Health Organization, Geneva, 2
nd
Edition.
Yang, Y., Pedersen, J.O., 1997. A comparative study on
feature selection in text categorization. In Proc. of the
14
th
International Conference on Machine Learning
(ICML-97), Nashville, TN, 1997, pp. 412–420.
AN INFORMATION FILTERING SYSTEM FOR E-HEALTH - The Health-on-Net Experience
215
