
IN SILICO STUDY OF EXPRESSION PROFILES CORRELATION
BETWEEN MICRORNAS AND CANCEROUS GENES
Ka-Lok Ng
*
and Chia-Wei Weng
Department of Bioinformatics, Asia University, 500 Lioufeng Road, Wufeng Shiang, Taichung, 41354, Taiwan
Keywords: MicroRNA, Oncogene, Tumor suppressor gene, Gene expression profile, Correlation coefficient.
Abstract: We investigate the possibility that microRNA can act as an oncogene or tumor suppressor gene.
Experimentally verified microRNA target genes information (TarBase) are integrated with microRNA and
mRNA expression data (NCI-60) to study this hypothesis, in which the Pearson correlation and Spearman
rank coefficients are used to quantify these relations for nine cancer types. Correlation coefficients with
negative values are used to filter out microRNA targets. Biological annotations of the targets are supplied by
using the TAG, GO and KEGG records. The above information are utilized to provide a platform in
identifying potential cancer related microRNAs. A web based interface is set up for information query and
data display.
1 INTRODUCTION
MicroRNAs (miRNAs) are a class of small non-
coding RNAs that bind to its target mRNA sequence
in the 3′-untranslated region (3′UTR), and induce
either translation repression or mRNA degradation.
Recent studies indicated that microRNA could
possibly play an important role in human cancer
where microRNA targets oncogene (OCG) or tumor
suppressor gene (TSG) to regulate the gene
expression (Zhang et al., 2007, He and Cao, 2007,
Wu and Hu, 2006, Garzon et al., 2006). When
microRNA plays an oncogenic role, it targets TSG
and leads to tumor formation. On the other hand, if
microRNA plays the tumor suppressor role, it would
target OCG and suppress tumor formation.
This work utilized the following databases; the
TarBase (Sethupathy et al., 2005), miRBase
(Griffiths-Jones et al., 2006) and NCI-60
(Shankavaram et al., 2007, Blower et al., 2007),
Online Mendelian Inheritance in Man (OMIM),
Tumor Associate Gene (TAG) (Chan, 2006) Gene
Ontology (Gene Ontology Consortiium, 2006), and
Kyoto Encyclopedia of Genes and Genomes (KEGG)
(Kanehisa et al., 2008) databases to set up a platform
for predicting human microRNA targeting cancerous
genes information. Table 1 states the general
information provided by the databases used in the
current study.
The platform mainly provides the following two
functionalities; (i) human microRNA target
information, and (ii) nine cancer types’ Pearson
correlation and Spearman rank coefficients of
microRNA and its target expression level for three
Affymetrix chips.
Table 1: General information provided by the databases
used in the current study.
Database General information provided
TarBase experimentally tested miRNA target genes
miRBase
information for precursor microRNAs,
mature microRNAs, FASTA sequences,
and their target genes.
NCI-60
human cancer cell lines mRNA and
miRNA expression data
OMIM human diseases genetic data
TAG OCG, TCG and cancer related genes
GO
Three types of gene annotations; i.e.
molecular function, biological
p
rocess and
subcellular localization
KEGG Metabolic pathways, disease pathways
141
Ng K. and Weng C. (2010).
IN SILICO STUDY OF EXPRESSION PROFILES CORRELATION BETWEEN MICRORNAS AND CANCEROUS GENES.
In Proceedings of the First International Conference on Bioinformatics, pages 141-146
DOI: 10.5220/0002728701410146
Copyright
c
SciTePress

2 MATERIALS AND METHODS
It is known that microRNA binds with mRNA and
can induce mRNA cleavage or inhibit translation. In
order to investigate the regulatory role of microRNA
in cancer diseases, we study the expression profiles
correlation between microRNA and its target genes,
in particular the OCG and TSG targets.
Figure 1 depicted the process flowchart for
identifying potential cancer related microRNAs. The
microRNA-target pairs information is obtained from
TarBase, whereas the expression profiles for
microRNAs and mRNAs are retrieved from the
NCI-60 dataset. Then, the expression profiles
correlation between microRNA and its target genes
are quantified by computing the correlation
coefficients. If the microRNA and its target gene are
direct interact, the correlation coefficient results
should reveal significant negative values.
MicroRNA target pairs with the correlation
coefficients below a given threshold are filtered for
further investigation. These pairs suggest a
regulatory relationship between the microRNAs and
their targets. The TAG dataset is used in order to sort
out the microRNA-OCG and microRNA-TSG pairs.
These pairs are further annotated by using the
OMIM, GO and KEGG biomedical terms. With
multiple biological annotations, i.e. disease type,
relevant biological function and pathway, for the
negative correlated pairs, this platform should
provide helpful guidance for investigating the role of
micorRNAs in tumor formation.
Figure 1: Process flowchart to identify potential cancer
related microRNAs based on quantifying the expression
profile correlation between microRNA and its target genes.
2.1 TarBase and miRBase Datasets
TarBase is a manually curated collection of
experimentally tested microRNA target genes. Each
experimentally validated target site is extracted from
the literatures. TarBase includes several species,
such as human, mouse, fruit fly, and worm,
microRNA target gene records.
There are many tools available for microRNA
target genes prediction, such as miRanda (Enright et
al., 2003), RNAhybrid (Kruger and Rehmsmeier,
2006), and TargetScans
(http://genes.mit.edu/tscan/targetscanS2005.html). A
major problem of microRNA target genes prediction
is that the prediction accuracy remains rather high,
there were reports indicated that the false positive
rate could be as high as 50% for human (John et al.,
2004), 24-39% and 22-31% when using miRanda
(Bentwich, 2005), and TargetScan (Bentwich, 2005)
respectively.
The primary goal of this work is to develop a
bioinformatics tool to investigate the possibility that
microRNA can act as an OCG or TSG. The main
advantage of using TarBase in constructing the
microRNA targeting information is that all the target
genes recorded by TarBase are experiment verified,
and TarBase provides their PubMed ID. From a
biologist point of view, experimental verified targets
imply higher confidence. If the miRNA:mRNA
targeting part is uncertain, then any further results
derived are doubtful. The TarBase version 5 dataset
from DIANA Lab. website is employed in the
present study. The miRBase database collects
information for precursor microRNAs, mature
microRNAs, FASTA sequences, and their target
genes. Currently the latest version of the miRBase
sequence database is 13.0, which includes
microRNA information across 103 species. In
version 13, a total of 706 Homo Sapiens mature
micorRNA entries are recorded.
2.2 Expression Datasets
In this study, we made use of the NCI-60 cancer cell
line mutation data to investigate the possibility that
microRNA can act as an OCG or TSG. This can be
achieved by calculating the correlation coefficient
between the expression levels of microRNAs and
their experimentally validated target genes.
The NCI-60 is a set of 60 human cancer cell lines
derived from diverse tissues. These cell lines include
nine tissues’ microRNA and mRNA expression
information, that is, breast cancer, central neural
system (CNS) cancer, colon cancer, leukemia,
melanoma, non-small cell lung cancer, ovarian
TarBase
NCI-60
Expression profile PCC, SRC calculation
PCC or SRC less
than -0.5
Integration of
OMIM, TAG,
KEGG, GO
Inferring potential cancer-related microRNAs
BIOINFORMATICS 2010 - International Conference on Bioinformatics
142
cancer, prostate cancer, and renal cancer,.
Four publicly available datasets of gene expression
profiles are selected in this study; including the
microRNA expression, and the Affymetrix U95(A-
E), U133A and U133B mRNA expression datasets.
Affymetrix mRNA expression datasets use three
types of normalization methods, that is, GCRMA,
MAS5 and RMA. Therefore, a total of ten datasets
are used, including one microRNA dataset and nine
Affymetrix RNA expression datasets. The NCI-60
website provides a tool, called CellMiner
(Shankavaram et al, 2009), to query those chip
datasets.
2.3 Tumor Associated Gene Database
The Tumor Associated Gene (TAG) database
presents information about cancer related genes. In
TAG, cancer related genes are classified into OCGs,
TSGs and tumor-associated genes. All genes in the
TAG are retrieved through text-mining approach
from the PubMed database. Currently, TAG
documented 519 genes, including 198 OCGs, 170
TSGs, and 151 genes related to oncogenesis. In
addition, more cancer related microRNA gene
information are obtained by using Pipeline Pilot
TM
,
which is a commercial bioinformatics text mining
package to do keywords search against PubMed. At
present, a total of 111 microRNAs are retrieved that
are related to certain types of cancers. These
microRNAs and their target records are stored in our
platform for further analysis.
2.4 OMIM, GO and KEGG Databases
Online Mendelian Inheritance in Man (OMIM) is a
compendium of human genes and genetic
phenotypes. It contains information on all known
mendelian disorders.
The GO database includes three structured
controlled vocabularies (ontology) that describe
gene products in terms of their associated biological
processes, cellular components and molecular
functions in a species-independent manner.
KEGG is short for Kyoto Encyclopedia of Genes
and Genomes which is a collection of manually
drawn pathway maps. In our study, we focused on
cancer related pathways information.
Investigation of cancer related pathway in
KEGG can help us determine the biological
functions of the target genes. We investigate which
cancer related pathways consist of the microRNA
target genes. By going through biological function
keywords search, a list of microRNA target genes
which participated in certain KEGG pathways are
obtained. Table 2 shows the cancer related pathways
which are processed in this study.
Table 2: Cancer related pathways which are processed in
this study.
KEGG ID Types of Cancer
hsa05210 Colorectal cancer
hsa05221 Acute myeloid leukemia
hsa05220 Chronic myeloid leukemia
hsa05218 Melanoma
hsa05211 Renal cell carcinoma
hsa05215 Prostate cancer
hsa05223 Non-small cell lung cancer
2.5 Preprocessing
The TarBase and NCI-60 datasets used different ID
formats for microRNAs, therefore, both sets of ID
are standardized using the miRBase IDs. We also
converted the mRNA gene IDs in TarBase and NCI-
60 Affymetrix RNA expression datasets IDs to
NCBI official symbols by using Gene Name Service
(Lin et al., 2007). Gene Name Service website
provides query services for 26 types of gene
identifiers of Homo sapiens genes. We obtained
44855, 20169 and 16441 entries for the U95(A-E),
U133A and U133B mRNA datasets after pre-
processing.
3 METHODS
3.1 Expression Profiles Correlation
For a given cancer tissue type, we calculated both
the Pearson correlation coefficient (PCC) and
Spearman rank coefficient (SRC),
ρ,
between the
expression level of a microRNA and it's target genes.
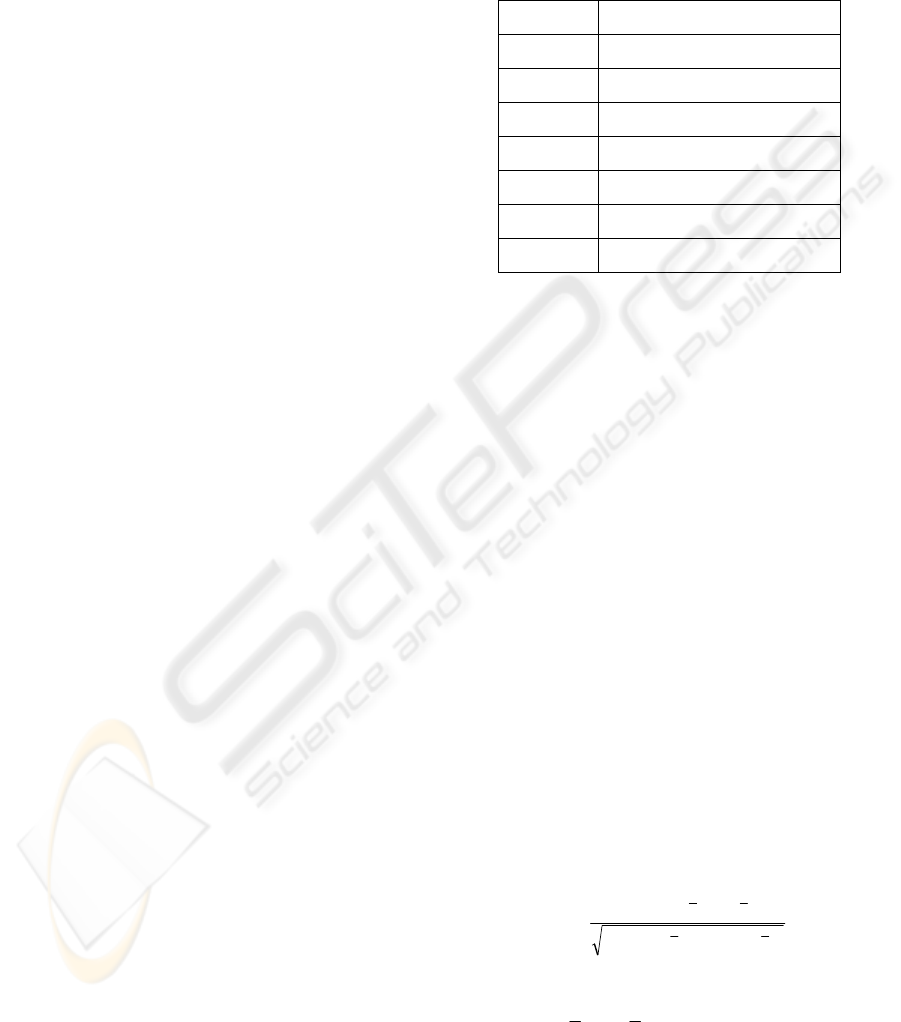
PCC or SRC is given by
∑∑
∑
==
=
−−
−−
=
n
i
n
i
ii
n
i
ii
yyxx
yyxx
11
22
1
)()(
))((
ρ
(1)
where x
i
and y
i
denote the expression intensity of
microRNA and the microRNA's target gene
respectively;
x
and
y
denote the mean expression
intensity of microRNA and the target gene
respectively; and n is the total number of the
IN SILICO STUDY OF EXPRESSION PROFILES CORRELATION BETWEEN MICRORNAS AND CANCEROUS
GENES
143
expression data entries. In case of SRC, the
expression intensity values of x
i
, y
i
,
x
and
y
are
replaced by their ranks.
The PCC for each microRNA and Affymetrix
RNA expression profile are computed for nine tissue
types. PCC takes a value between -1 and +1. One of
the troubles with quantifying the strength of
correlation by PCC is that it is susceptible to being
skewed by outliers. Outliers that is a single data
point can result in two genes appearing to be
correlated, even when all the other data points not.
SRC is a non-parametric statistical method that is
robust to outliers. It can ignore the magnitude of the
changes. The idea of SRC is to transform the
original values into ranks, and then to compute the
correlation between the series of ranks.
Gene expression values of microRNA and
mRNA in the same tissue type are ordered in
ascending order, the lowest value is assigned to rank
one. In case of ties mid-rank is assigned, as for
example, when both values are ranked five, a rank of
5.5 is assigned. After ranking the expression profiles
of microRNA and mRNA for a particular tissue,
SRC can be calculated by Eq. (1), with the ranks and
the average value of ranks of microRNA and mRNA
are used instead of the expression intensity and
average expression intensity. The SRC also takes a
value between -1 and +1.
4 RESULT
Both the PCC and SRC of microRNA expression
levels and their targeting mRNA expression levels
for nine types of cancer tissues are computed.
For example, microRNA hsa-miR-16 targets the
breast cancer gene, BCL2, have PCC less than -0.7
for the three Affymetrix datasets with three different
normalization methods for each chip. The results are
reported in Table 3. We can understand negative
PCC (SRC) based on the following reasoning. It is
known that microRNA is able to repress and/or
cleavage mRNA by incomplete or complete
complementary binding with the mRNA. If a
microRNA and its target gene is directly interacting,
the result of PCC (SRC) of their expression profiles
should reveal negative correlation. Table 3 list the
U95 results which suggested that microRNA, hsa-
miR-16, can possibly play a role in regulating the
cancer gene BCL2.
Downstream cancer targets are easily obtained by
cross-referencing the target gene results with the
TAG dataset. The results of cancerous genes found
in both TarBase and TAG are listed in Table 4.
Table 3: PCC of the expression profile for hsa-miR-16 and
BCL2 in the breast tissue.
Dataset Normalization PCC
U95(A-E) GCRMA -0.918
MAS5 -0.949
RMA -0.718
Table 4: MicroRNA target genes which are corresponded
with TAG entries.
4.1 Evaluation of Results by OMIM
Disease disorder keywords provided by the OMIM,
is compared with the negative (under or equal to -0.5)
PCC results. It is found that 82 entries involved in
disease disorder. Among these 82 entries, 19 are
cancer genes. In these cancer genes, only 17 entries
belong to the nine cancer types. Table 5 presents
five of the 17 OMIM entries.
Table 5: The 17 OMIM entries with negative PCC ( -
0.5) and their corresponding cancerous types.
OMIM ID Gene Symbol Cancerous Types
605882 BACH1 Breast
151430 BCL21 Leukemia
168461 CCND1 Colon
123829 CDK41 Melanoma
600160 CDKN2A CNS||Melanoma
For a given Affymetrix chip, it is found that the
PCC score is independent of the normalization
methods. Similarly, for a given normalization
method, the PCC score is also rather independent of
the chip type. Furthermore, it is also found that
target results (microRNA:mRNA) obtained by PCC
versus PCC and SRC are rather consistent. Table 4
reports the results for the U95(A-E) chip with the
GCRMA normalization method.
According to Table 6, four pairs of negative
microRNA:mRNA are found belonging to leukemia,
and one pair belongs to lung cancer. Among these
five pairs, only hsa-mir-370 targets an OCG, i.e.
MAP3K8. Besides the OMIM recorded data in
Table 4, we also identified five (in Italic) microRNA
Cancer Gene Gene Symbols
OCG
AXL, BCL2, CCND1, CDK4, CTGF,
ESR1, FGF20, KIT, MAP3K8, MYB,
MYBL1, MYCN, RYK, TEAD1
TSG
CAV1, CDKN1A, CDKN1C, CDKN2A,
HTATIP2, MXI1, NF2, PTEN, PTPN12,
RB1, SERPINB5, TGFBR2
BIOINFORMATICS 2010 - International Conference on Bioinformatics
144
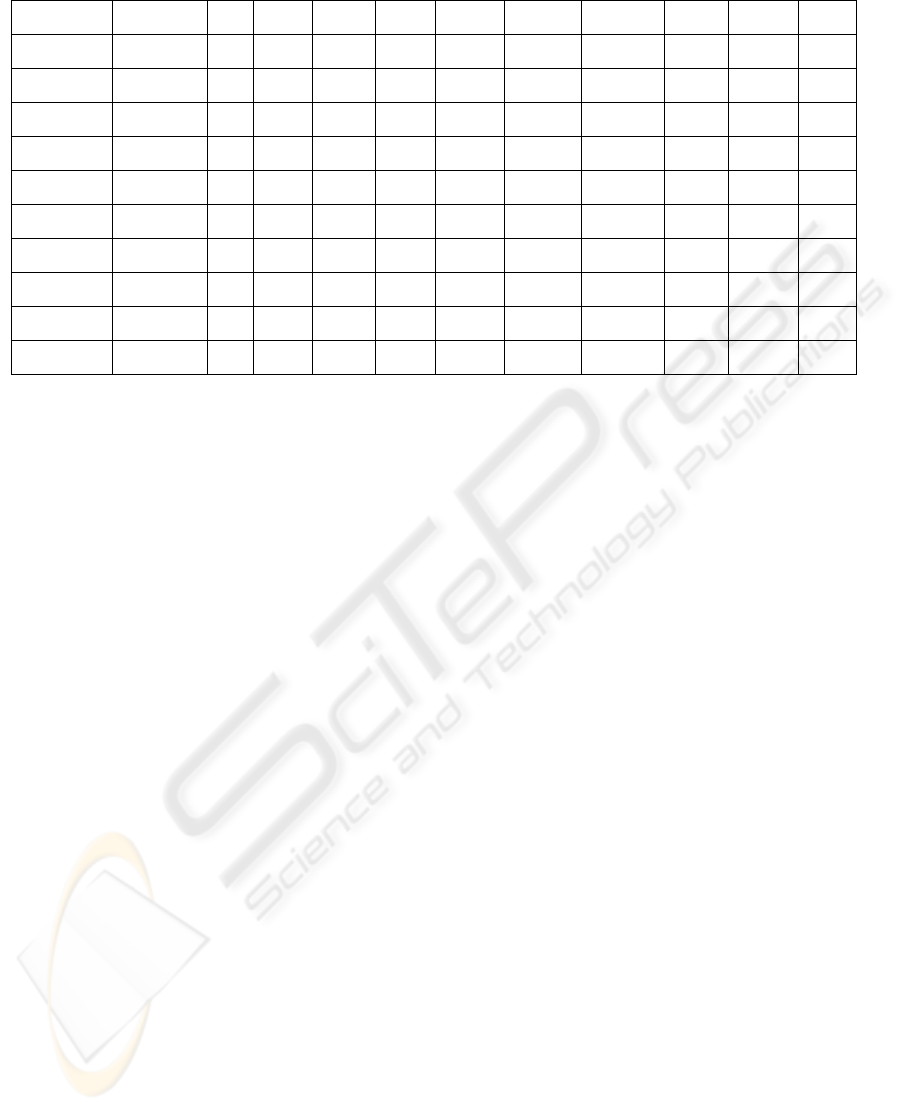
Table 6: OMIM evaluation results of both PCC and SRC under or equal to -0.5 for GCRMA-U95(A-E) dataset.
Bold font denotes the OMIM matching data.
Italic and bold font denotes that half of the microRNA probes’
PCC and SRC are ≦-0.5. Inside the box, the numerator of the fraction denotes the number of times where both of PCC and
SRC are less than -0.5.
The denominator represents the total number of PCC values calculated (i.e. the number of microRNA probes times the
number of mRNA probes). Inside the square bracket, zero ([0]) implies no matching with the OMIM data and [*] denotes
matching with the OMIM data.
targets in which half of the probes’ PCC and SRC
are under or equal to -0.5 in prostate cancer. Among
these pairs, one of the target is a TSG, i.e.
CDKN2A, and two targets are OCGs, i.e. CCND1
and MAP3K8.
4.2 Evaluation of Results by KEGG
The KEGG pathways are compared with the PCC
results. A total of 26 pairs (with an asterisk) of
negative correlated microRNA and its target genes
matched with KEGG data for four cancer types,
which are colon, leukemia, melanoma and prostate.
Among these KEGG matched pairs, twelve pairs
(with an asterisk) have both the PCC and SRC under
or equal to -0.5 in more than half of the probes. In
these twelve pairs, two target genes are TSG, i.e.
CDKN1A and PTEN, and one gene is an OCG, i.e.
CCND1.
In OMIM and KEGG evaluation results, two
pairs of microRNA:mRNA, i.e. hsa-mir-24-
1:CDKN2A and hsa-mir-24-2:CDKN2A, both
present significant negative correlation in prostate
cancer, with the correlation coefficients under or
equal to -0.5 in more than half of the probes. At
present, it is still unclear whether the hsa-mir-24-2
and CDKN2A has any regulatory relationship in
prostate cancer. These predicted negative correlated
microRNA:mRNA pairs maybe subjected to further
investigation in order to identify the exact regulatory
situations in prostate cancer.
According to OMIM (Table 6) and KEGG
evaluation results, one pair presents negative
correlation in leukemia which is hsa-mir-
34a:CCND1, but the PCC and SRC scores don’t
present in more than half of the probes are under or
equal to -0.5. Although the evidence is not strong,
but it suggests that hsa-mir-34a may also be a
potential regulator of CCND1 in leukemia.
Final, we set up a web based service to provide
the computed results. The web site is availabled at
http://ppi.bioinfo.asia.edu.tw/mirna_target/index.html.
5 CONCLUSIONS
Recent studies indicate that microRNA could
possibly play an important role in human cancer,
where microRNA targets TSG or doesn’t target
OCG. Experimentally verified microRNA targeted
genes information (TarBase) are integrated with
microRNA and mRNA expression data (NCI-60) to
study this hypothesis, in which two correlation
coefficients, PCC and SRC, are used to quantify the
correlation between microRNA and its targets
expression profiles. The predicted results are
evaluated with reference to the OMIM and KEGG
data. It is found that the obtained results are rather
Precursor Target Gene TAG Breast CNS Colon Lung Leukemia Melanoma Ovarian Prostate Renal
hsa-let-7b CCND1 OCG 0/3[0] 0/3[0] 0/3[0] 0/3[0] 0/3[0] 0/3[0] 0/3[0] 0/3[0] 1/3[0]
hsa-mir-155 BACH1 - 0/1[0] 0/1[0] 0/1[0] 0/1[0] 0/1[0] 0/1[0] 0/1[0] 0/1[0] 0/1[0]
hsa-mir-23b NOTCH1 - 1/4[0] 0/4[0] 0/4[0] 0/4[0] 1/4[*] 0/4[0] 0/4[0] 1/4[0] 0/4[0]
hsa-mir-24-1 NOTCH1 - 2/8[0] 0/8[0] 1/8[0] 0/8[0] 2/8[*] 0/8[0] 1/8[0] 2/8[0] 0/8[0]
hsa-mir-24-2 NOTCH1 - 1/4[0] 0/4[0] 0/4[0] 0/4[0] 1/4[*] 0/4[0] 0/4[0] 1/4[0] 0/4[0]
hsa-mir-24-1 CDKN2A TSG 0/8[0] 0/8[0] 2/8[0] 0/8[0] 0/8[0] 0/8[0] 2/8[0] 6/8[0] 1/8[0]
hsa-mir-24-2 CDKN2A TSG 0/4[0] 0/4[0] 0/4[0] 0/4[0] 0/4[0] 0/4[0] 1/4[0] 3/4[0] 0/4[0]
hsa-mir-27b NOTCH1 - 1/4[0] 0/4[0] 0/4[0] 0/4[0] 1/4[*] 0/4[0] 1/4[0] 3/4[0] 0/4[0]
hsa-mir-34a CCND1 OCG 0/6[0] 0/6[0] 0/6[0] 0/6[0] 1/6[0] 0/6[0] 0/6[0] 3/6[0] 0/6[0]
hsa-mir-370 MAP3K8 OCG 0/14[0] 1/14[0] 0/14[0] 1/14[*] 0/14[0] 0/14[0] 2/14[0] 8/14[0] 0/14[0]
IN SILICO STUDY OF EXPRESSION PROFILES CORRELATION BETWEEN MICRORNAS AND CANCEROUS
GENES
145

independent of the chip types and the normalization
methods too.
In the OMIM evaluation with both PCC and SRC
less than or equals to -0.5, five pairs of negative
correlated microRNA and its target genes matched
with OMIM records, in which four of them belong
to leukemia and the rest one is lung cancer. In these
five pairs, only one of them is an OCG, i.e.
MAP3K8. Besides, we also got five pairs of
significantly negative correlated microRNA and its
target in prostate cancer in which both of PCC and
SRC are under or equals to -0.5. Among these five
pairs, only one gene is a TSG, i.e. CDKN2A, and
only two genes are OCGs, i.e. CCND1 and
MAP3K8. These five pairs can be browsed in Table
5 in which they are denoted with italic and bold font.
Similar conclusions are obtained for the KEGG
evaluation.
Given that more than half of the probes’
correlation coefficients are negative correlated, we
identified certain putative pairs of microRNA and its
cancer related targets in different cancer types, such
as, hsa-mir-24-1:CDKN2A and hsa-mir-24-
2:CDKN2A in prostate cancer and hsa-mir-
19a:PTEN in both leukemia and prostate cancer.
It is suggested that those negative correlated pairs of
microRNA and target can be subjected to further
investigation, such as performing in vivo
experiments to valid the hypothesis that microRNA
could possibly play an important role in human
cancer.
ACKNOWLEDGEMENTS
K-L Ng work is supported by the National Science
Council of R.O.C. under the grant of NSC 98-2221-
E-468-013.
REFERENCES
Bentwich I. 2005. Prediction and validation of
microRNAs and their targets. FEBS Lett., 579, 5904.
Blower P.E., Verducci JS, Lin S, Zhou J, Chung JH, Dai Z,
Liu CG, Reinhold W, Lorenzi PL, Kaldjian EP, Croce
CM, Weinstein JN, Sadee W., 2007. MicroRNA
expression profiles for the NCI-60 cancer cell panel,
Mol. Cancer Ther., 6, 1483-1491.
Chan Hsiang-Han 2006. Identification of novel tumor-
associated gene (TAG) by bioinformatics analysis.
MSc. Thesis, Institute of Molecular Medicine,
National Cheng Kung University, Taiwan.
Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C.,
Marks, D.S., 2003. MicroRNA targets in Drosophila.
Genome Biology, 5(1):R1.
Garzon Ramiro, Fabbri Muller, Cimmino Amelia, Calin
George A. and Croce Carlo M., 2006. MicroRNA
expression and function in cancer. Trends in
Molecular Medicine, 12, 580-588.
Gene Ontology Consortium 2006. The Gene Ontology
(GO) project in 2006. Nucl. Acids Res, 34, D322–326.
Griffiths-Jones S., Grocock Russell J., van Dongen Stijn,
Bateman Alex, Enright Anton J., 2006. miRBase:
microRNA sequences, targets and gene nomenclature.
Nucl. Acids Research, 34, 140-144.
He Xiaoting, Cao Xiufeng 2007. MicroRNA and
esophageal carcinoma. J.N.M.U., 21, 201-206.
John B., Enright A.J., Aravin A., Tuschl T., Sander C.,
Marks D.S., 2004. Human MicroRNA targets. PLoS
Biol. 2(11), e363.
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M.,
Itoh M., Katayama T., Kawashima S., Okuda S.,
Tokimatsu T., and Yamanishi Y., 2008. KEGG for
linking genomes to life and the environment. Nucl.
Acids Res., 36, 480-484
Kruger Jan and Rehmsmeier Marc 2006. RNAhybrid:
miRNA target prediction easy, fast and flexible,
Nucleic Acids Research, 34, 451-454.
Lin Kuan-Ting, Liu Chia-Hung, Chiou Jen-Jie, Tseng
Wen-Hsien, Lin Kuang-Lung, Hsu Chun-Nan 2007.
Gene Name Service: No-Nonsense Alias Resolution
Service for Homo Sapiens Genes. Proceedings of the
2007 IEEE/WIC/ACM International Conferences on
Web Intelligence and Intelligent Agent Technology-
Workshops, 185-188.
Sethupathy P., Corda B., Hatzigeorgiou A. 2005 TarBase:
A comprehensive database of experimentally
supported animal microRNA targets. RNA, 12, 192-
197.
Shankavaram U.T., Uma T., Reinhold William C.,
Nishizuka Satoshi, Major Sylvia, Morita Daisaku,
Chary Krishna K., Reimers Mark A., Scherf Uwe,
Kahn Ari, Dolginow Douglas, Cossman Jeffrey,
Kaldjian Eric P., Scudiero Dominic A., Petricoin
Emanuel, Liotta Lance, Lee Jae K., Weinsteinu John
N, 2007. Transcript and protein expression profiles of
the NCI-60 cancer cell panel: an integromic
microarray study. Molecular Cancer Therapeutics, 6,
820-832.
Shankavaram U.T., Varma S, Kane D, Sunshine M, Chary
KK, Reinhold WC, Pommier Y, Weinstein JN., 2009.
CellMiner: a relational database and query tool for the
NCI-60 cancer cell lines. BMC Genomics,10, 277..
Wu Dan and Hu Lan 2006. Micro-RNA: A New Kind of
Gene Regulators. Agricultural Sci. in China, 5, 77-80.
Zhang Baohong, Pan Xiaoping, Cobb George P.,
Anderson Todd A., 2007. microRNAs as oncogenes
and tumor suppressors. Develop. Biol, 302, 1-12.
BIOINFORMATICS 2010 - International Conference on Bioinformatics
146
