
SELECTING GENES FROM GENE EXPRESSION DATA BY
USING AN ENHANCEMENT OF BINARY PARTICLE SWARM
OPTIMIZATION FOR CANCER CLASSIFICATION
Mohd Saberi Mohamad
1,2
, Sigeru Omatu
1
, Michifumi Yoshioka
1
and Safaai Deris
2
1
Department of Computer Science and Intelligent Systems, Osaka Prefecture University, Sakai, Osaka 599-8531, Japan
2
Department of Software Engineering, Universiti Teknologi Malaysia, 81310 Skudai, Johore, Malaysia
Keywords: Binary particle swarm optimization, Gene selection, Gene expression data, Cancer classification.
Abstract: In order to select a small subset of informative genes from gene expression data for cancer classification,
recently, many researchers are analyzing gene expression data using various computational intelligence
methods. However, due to the small number of samples compared to the huge number of genes (high-
dimension), irrelevant genes, and noisy genes, many of the computational methods face difficulties to select
the small subset. Thus, we propose an enhancement of binary particle swarm optimization to select a small
subset of informative genes that is relevant for classifying cancer samples more accurately. In this proposed
method, three approaches have been introduced to increase the probability of bits in particle’s positions to
be zero. By performing experiments on three different gene expression data sets, we have found that the
performance of the proposed method is superior to other previous related works, including the conventional
version of binary particle swarm optimization (BPSO) in terms of classification accuracy and the number of
selected genes. The proposed method also produces lower running times compared to BPSO.
1 INTRODUCTION
Recent advances in microarray technology allow
scientists to measure the expression levels of
thousands of genes simultaneously in biological
organisms and have made it possible to create
databases of cancerous tissues. It finally produces
gene expression data that contain useful information
of genomic, diagnostic, and prognostic for
researchers (Knudsen, 2002). Thus, there is a need to
select informative genes that contribute to a
cancerous state. However, the gene selection process
poses a major challenge because of the following
characteristics of gene expression data: the huge
number of genes compared to the small number of
samples (high-dimensional data), irrelevant genes,
and noisy data. To overcome this challenge, a gene
selection method is usually used to select a subset of
informative genes that maximizes classifier’s ability
to classify samples more accurately (Mohamad et
al., 2009). The advantages of gene selection has
been reported in Mohamad et al. (2009).
Recently, several gene selection methods based
on particle swarm optimization (PSO) have been
proposed to select informative genes from gene
expression data (Shen et al., 2008; Chuang et al.,
2008; Li et al., 2008). PSO is a new evolutionary
technique proposed by Kennedy and Eberhart
(1995). It is motivated from the simulation of social
behavior of organisms such as bird flocking and fish
schooling. Shen et al. (2008) have proposed a hybrid
of PSO and tabu search approaches for gene
selection. However, the results obtained by using the
hybrid method are less meaningful since the
application of tabu approaches in PSO is unable to
search a near-optimal solution in search spaces.
Next, an improved binary PSO have been proposed
by Chuang et al. (2008). This approach produced
100% classification accuracy in many data sets, but
it used a high number of selected genes (large gene
subset) to achieve the high accuracy. It uses the high
number because of the global best particle is reset to
zero position when its fitness values do not change
after three consecutive iterations. After that, Li et al.
(2008) have introduced a hybrid of PSO and genetic
algorithms (GA) for the same purpose.
Unfortunately, the accuracy result is still not high
and many genes are selected for cancer classification
82
Saberi Mohamad M., Omatu S., Yoshioka M. and Deris S. (2010).
SELECTING GENES FROM GENE EXPRESSION DATA BY USING AN ENHANCEMENT OF BINARY PARTICLE SWARM OPTIMIZATION FOR
CANCER CLASSIFICATION.
In Proceedings of the 2nd International Conference on Agents and Artificial Intelligence - Artificial Intelligence, pages 82-89
DOI: 10.5220/0002732300820089
Copyright
c
SciTePress

since there are no direct probability relations
between GA and PSO. Generally, the PSO-based
methods (Shen et al., 2008; Chuang et al., 2008; Li
et al., 2008) are intractable to efficiently produce a
small (near-optimal) subset of informative genes for
high classification accuracy. This is mainly because
the total number of genes in gene expression data is
too large (high-dimensional data).
Therefore, we propose an enhancement of binary
PSO (EPSO) to select a small (near-optimal) subset
of informative genes that is most relevant for
classifying cancer classes more accurately. In order
to test the effectiveness of our proposed method, we
apply EPSO to three gene expression data sets,
including binary-classes and multi-classes data sets.
This paper is organized as follows. In Section 2,
we briefly describe the conventional version of
binary PSO and EPSO. Section 3 presents data sets
used and experimental results. Section 4 summarizes
this paper by providing its main conclusions and
addresses future developments.
2 METHODS
2.1 The Conventional Version of
Binary PSO (BPSO)
BPSO is initialized with a population of particles. At
each iteration, all particles move in a problem space
to find the optimal solution. A particle represents a
potential solution in an n-dimensional space
(Kennedy and Eberhart, 1997). Each particle has
position and velocity vectors for directing its
movement. The position vector and velocity vector
of the ith particle in the n-dimension can be
represented as
12
(, ,..., )
n
iii i
X
xx x=
and
12
( , ,..., )
n
iii i
Vvv v= , respectively, where {0,1};
d
i
x ∈
i=1,2,..m (m is the total number of particles); and
d=1,2,..n (n is the dimension of data).
d
i
v is a real
number for the d-th dimension of the particle i,
where the maximum
d
i
v ,
max
(1 / 3) .Vn=×
In gene selection, the vector of particle positions
is represented by a binary bit string of length n,
where n is the total number of genes. Each position
vector
()
i
X
denotes a gene subset. If the value of
the bit is 1, it means that the corresponding gene is
selected. Otherwise, the value of 0 means that the
corresponding gene is not selected. Each particle in
the t-th iteration updates its own position and
velocity according to the following equations:
11
22
(1)()() ()( ()
()) () ( () ())
dddd
ii i
dd dd
ii
vt wt vt crt pbestt
x t c r t gbest t x t
+= × + ×
−+ × −
(1)
(1)
1
((1))
1
d
i
d
i
vt
Sig v t
e
−+
+=
+
(2)
if
3
(( 1)) (),
dd
i
Sig v t r t+> then (1)1;
d
i
xt+=
else
(1)0
d
i
xt
+
=
(3)
where
1
c and
2
c are the acceleration constants in
the interval [0,2].
123
(), (), ()~ (0,1)
ddd
rtrtrt U are
random values in the range [0,1] that sampled from a
uniform distribution.
12
() ( (), (),..., ())
n
iii i
Pbest t pbest t pbest t pbest t= and
12
() ( (), (),..., ())
n
Gbest t gbest t gbest t gbest t= represent
the best previous position of the ith particle and the
global best position of the swarm (all particles),
respectively. They are assessed base on a fitness
function.
(( 1))
d
i
Sig v t
+
is a sigmoid function where
((1))[0,1].
d
i
Sig v t +∈
()wt
is an inertia weight and
initialized with 1.4. It is updated as follows:
(() 0.4)( ())
(1)
(0.4)
wt MAXITER Itert
wt
MAXITER
−× −
+=
+
(4)
where
M
AXITER
is the maximum iteration
(generation) and
()Iter t is the current iteration.
2.1.1 Investigating the Drawbacks of BPSO
and Previous PSO-based Methods
Before attempting to propose EPSO, it would be
prudent to find the limitations of BPSO and previous
PSO-based methods (Shen et al., 2008; Chuang et
al., 2008; Li et al., 2008). This subsection
investigates theoretically the limitations by
analyzing Eq. 2 and Eq. 3. These equations are
analyzed because they are most important equations
for genes selection in binary spaces. Both the
equations are also implemented in BPSO and the
PSO-based methods.
The sigmoid function (Eq. 2) represents a
probability for
()
d
i
x
t to be 0 or 1 ( (()0)
d
i
Px t
=
or
(()1)
d
i
Px t
=
). For example,
if
() 0,
d
i
vt
=
then (()0)0.5
d
i
Sig v t == and
( ()0)0.5.
d
i
Px t ==
if
() 0,
d
i
vt
<
then
(()0)0.5
d
i
Sig v t <<
and
( ()0)0.5.
d
i
Px t =>
SELECTING GENES FROM GENE EXPRESSION DATA BY USING AN ENHANCEMENT OF BINARY PARTICLE
SWARM OPTIMIZATION FOR CANCER CLASSIFICATION
83

if
() 0,
d
i
vt>
then
(()0)0.5
d
i
Sig v t >>
and
(()0)0.5.
d
i
Px t =<
Also note that
).1)((1)0)(( =−== txPtxP
d
i
d
i
From the analysis,
we conclude that
(()0) (()1)0.5
dd
ii
Px t Px t== ==
because Eq. 2 is a standard sigmoid function without
any constraint and no modification. Hence, by using
this standard sigmoid function in high-dimensional
spaces (gene expression data), it only reduces the
number of genes to about half of the total number of
genes. This is reported and proved in the section of
experimental results. Therefore, Eq. 2 and Eq.3 are
potentially being the drawbacks of BPSO and the
previous PSO-based methods in selecting a small
number of genes for producing a near-optimal
(small) subset of genes from gene expression data.
2.2 An Enhancement of Binary PSO
(EPSO)
Almost all previous works of gene expression data
researches have selected a subset of genes to obtain
excellent cancer classification. Therefore, in this
article, we propose EPSO for selecting a near-
optimal (small) subset of genes. It is proposed to
overcome the limitations of BPSO and previous
PSO-based methods (Shen et al., 2008; Chuang et
al., 2008, Li et al., 2008). EPSO in our work differs
from BPSO and the PSO-based methods on three
parts: 1) we introduce a scalar quantity that called
particles’ speed
()
s
; 2) we propose a rule for
updating
(1)
d
i
xt+ ; 3) we modify the existing
sigmoid function, whereas BPSO and the PSO-based
methods have used the original rule (Eq. 3) and the
standard sigmoid function (Eq.2), and no particles’
speed implementation. The particles’ speed, rule,
and sigmoid function are introduced in order to:
increase the probability of
(1)0
d
i
xt
+
=
(( ( 1) 0))
d
i
Px t+= .
reduce the probability of (1)1
d
i
xt
+
=
(( ( 1) 1))
d
i
Px t+=
.
The increased and decreased probability values
cause a small number of genes are selected and
grouped into a gene subset.
(1)1
d
i
xt+= means that
the corresponding gene is selected. Otherwise,
(1)0
d
i
xt+= represents that the corresponding gene
is not selected.
Definition 1.
i
s
is a speed or length or magnitude of
i
V
for the particle i. In a real Euclidean space
n
ℜ
,
where
ℜ
denotes the field of real numbers, and n is
the dimension of
ℜ
,
i
s
can be derived by the
Euclidean norm as follows:
() ( ) ( )
22 2
12
...
n
ii i i i
sV v v v== + ++
(5)
Therefore, the following properties of
i
s
are crucial:
non-negativity:
0;
i
s ≥
definiteness:
0
i
s
=
if and only if 0;
i
V =
homogeneity:
iii
VVs
α
αα
== where 0;
α
≥
the triangle inequality:
11ii i i
VV V V
++
+≤+
where
ii
Vs
=
and
11ii
Vs
++
=
The particles’ speed, rule, and sigmoid function are
proposed as follows:
11
22
(1) () () () ( ()
()) () ( () ())
ii i
ii
s
twtstcrtdistPbestt
Xt crt distGbestt Xt
+= × + ×
−+ × −
(6)
5( 1)
1
(( 1))
1
i
i
st
Sig s t
e
−+
+=
+
subject to
(1)0
i
st
+
≥
(7)
if
3
(( 1)) (),
d
i
Sig s t r t+> then (1)0;
d
i
xt+=
else
(1)1
d
i
xt
+
=
(8)
where
(1)
i
st
+
represents the speed of the particle i
for the t+1 iteration, whereas in BPSO and previous
PSO-based methods (Eq. 1, Eq. 2, and Eq. 3),
(1)
d
i
vt
+
represents a single element of a particle
velocity vector for the particle i. In EPSO, Eq. 6, Eq.
7, and Eq. 8 are used to replace Eq. 1, Eq. 2, and Eq.
3, respectively.
(1)
i
st
+
is the rate at which the
particle i changes its position. Based on Definition 1,
the most important property of
(1)
i
st+ is
(1)0.
i
st
+
≥
Hence,
(1)
i
st
+
is used instead of
(1)
d
i
vt
+
so that its positive value can increase
((1)0).
d
i
Px t+=
In Eq. 6, the calculation for updating
(1)
i
st
+
is
mainly based on the distance between
()
i
Pbest t
and
()
i
X
t ( ( () ())),
ii
dist Pbest t X t− and the distance
between
()Gbest t and ()
i
X
t ( ( ( ) ( ))),
i
dist Gbest t X t−
whereas the original formula (Eq. 1) is used to
calculate
(1)
d
i
vt
+
and it is essentially based on the
ICAART 2010 - 2nd International Conference on Agents and Artificial Intelligence
84
()
1
(())
1
d
i
d
i
vt
Sig v t
e
−
=
+
0
1
0.5
0
(()1)
d
i
Px t
=
(()0)
d
i
Px t =
()
d
i
vt
(())
d
i
Sig v t
2 -2
b
)
0
1
0.5
0
2 -2
a
)
5()
1
(())
1
i
i
s
t
Sig s t
e
−
=
+
(()0)
d
i
Px t =
(()1)
d
i
Px t =
()
i
s
t
The area of
unsatisfied
() 0
i
st≥
(())
i
Sig s t
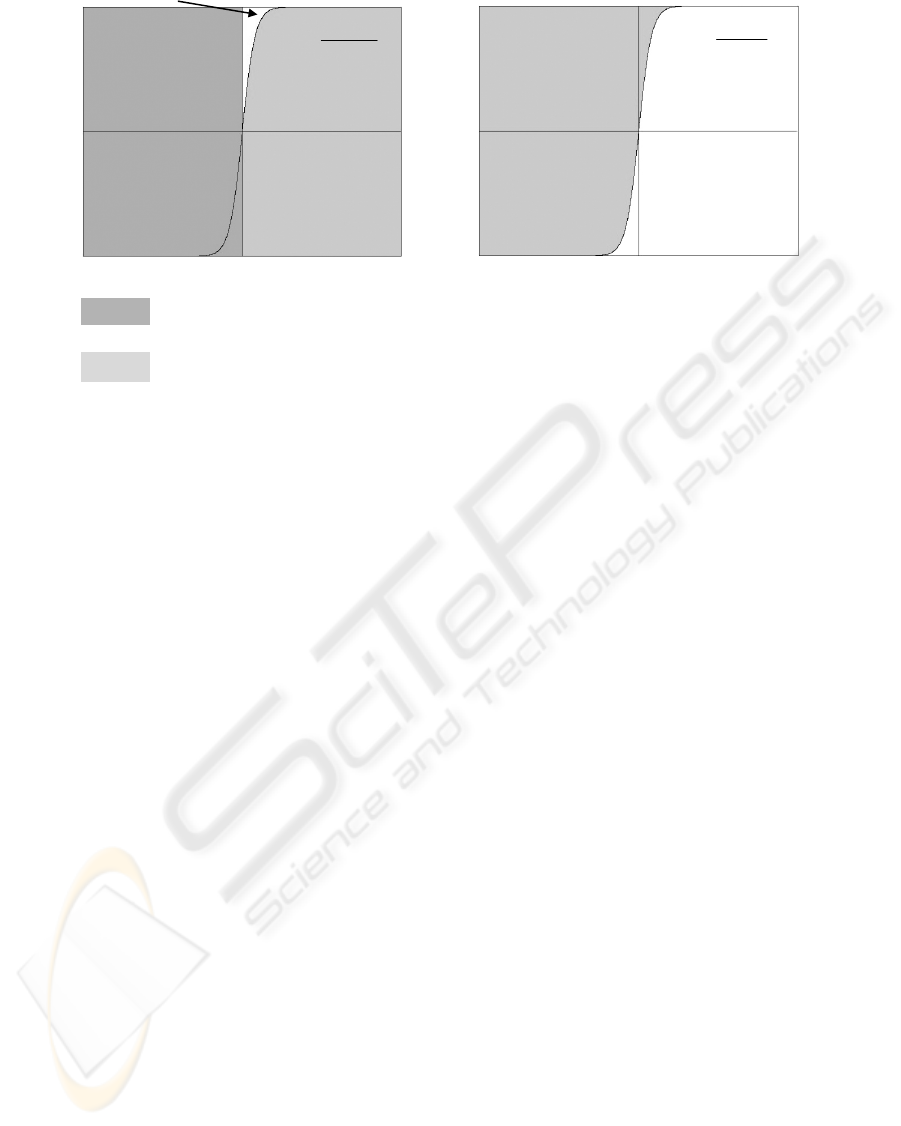
Figure 1: The areas of unsatisfied
() 0,
i
st≥
(()0)
d
i
Px t
=
and
(()1)
d
i
Px t
=
in a) EPSO; b) BPSO.
difference between ()
d
Gbest t and ().
d
i
x
t The
distances are used in the calculation for updating
(1)
i
st+ in order to always satisfy the property of
(1),
i
st+
namely
(( 1) 0)
i
st+≥
and finally increase
((1)0).
d
i
Px t+= Subsection 2.2.1 explains how to
calculate the distance between two positions of two
particles, e.g.,
( ( ) ( )).
i
dist Gbest t X t−
Theorem 1. Equations (6-8) and
() 0
i
st≥
increase
(()0)
d
i
Px t = because the minimum value for
(()0)
d
i
Px t = is 0.5 when () 0
i
st
=
(min ( ( ) 0) 0.5).
d
i
Px t =≥
Meanwhile, they decrease the
maximum value for
(()1)
d
i
Px t = to 0.5
(max ( ( ) 1) 0.5).
d
i
Px t =≤ Therefore, if () 0,
i
st> then
(()0) 0.5
d
i
Px t =>>
and
(()1) 0.5.
d
i
Px t =<<
Proof.
()⇒ Figure 1 shows that a) Equations (6-8)
and
() 0
i
st≥ in EPSO increase (()0);
d
i
Px t = b)
Equations (1-3) in BPSO yield
( ( ) 0) ( ( ) 1) 0.5.
dd
ii
Px t Px t== == For example, the
calculations for
(()0)
d
i
Px t = and (()1)
d
i
Px t = in Fig.
2(a) are shown as follows:
if
() 1,
i
st= then ( ( ) 0) 0.993307
d
i
Px t == and
( ( ) 1) 1 ( ( ) 0) 0.006693.
dd
ii
Px t Px t==− = =
if () 2,
i
st= then ( ( ) 0) 0.999955
d
i
Px t == and
( ( ) 1) 1 ( ( ) 0) 0.000045.
dd
ii
Px t Px t==− = =
Moreover, the modified sigmoid function (Eq. 7)
generates higher
(()0)
d
i
Px t = compared to the
standard sigmoid function (Eq. 2). For example, the
calculations for
(()0)
d
i
Px t
=
and (()1)
d
i
Px t = in Fig.
2(b) are shown as follows:
if
() 1,
i
st
=
then ( ( ) 0) 0.731059
d
i
Px t == and
( ( ) 1) 1 ( ( ) 0) 0.268941.
dd
ii
Px t Px t==− = =
if
() 2,
i
st
=
then ( ( ) 0) 0.880797
d
i
Px t == and
( ( ) 1) 1 ( ( ) 0) 0.119203.
dd
ii
Px t Px t==− = =
The high probability of
() 0
d
i
xt= (( () 0))
d
i
Px t =
causes a small number of genes are selected in order
to produce a near-optimal (small) gene subset from
high-dimensional data (gene expression data).
Hence, EPSO is proposed to overcome the
limitations of BPSO and the previous PSO-based
methods, and finally produce a small subset of
informative genes.
2.2.1 The Calculation of the Distance of Two
Particles’ Positions
The number of different bits between two particles
relates to the difference between their positions. For
example,
( ) [0011101000]Gbest t
=
and
( ) [1110110100].
i
Xt
=
The difference between
()Gbest t and
()
i
X
t
is [ 1 1010 11 100].
−
−−− The value
of 1 indicates that compared with the best position,
this bit (gene) should be selected, but it is not
selected, which may decrease classification quality
Legends:
The area of unsatisfied () 0.
i
st≥
The area of
(()0).
d
i
Px t =
The area of (()1).
d
i
Px t =
SELECTING GENES FROM GENE EXPRESSION DATA BY USING AN ENHANCEMENT OF BINARY PARTICLE
SWARM OPTIMIZATION FOR CANCER CLASSIFICATION
85

and lead to a lower fitness value. In contrast, a value
of -1 indicates that, compared with the best position,
this bit should not be selected, but it is selected. The
selection of irrelevant genes makes the length of the
subset longer and leads to a lower fitness value.
Assume that the number of 1 is
,a whereas the
number of -1 is
.b We use the absolute value of
ab− (| |)ab− to express the distance between two
positions. In this example, the distance between
()Gbest t and
()
i
X
t
is
(()())|||24|2.
i
dist Gbest t X t a b− =−=−=
2.2.2 Fitness Functions
The fitness value of a particle (a gene subset) is
calculated as follows:
12
() ()(( ())/)
ii i
fitness X w A X w n R X n=× + −
(9)
in which
() 0,1
i
AX ∈
⎡⎤
⎣⎦
is leave-one-out-cross-
validation (LOOCV) classification accuracy that
uses the only genes in a gene subset
().
i
X
This
accuracy is provided by support vector machine
classifiers (SVM).
()
i
R
X is the number of selected
genes in
.
i
X
n is the total number of genes for each
sample.
1
w and
2
w are two priority weights
corresponding to the importance of accuracy and the
number of selected genes, respectively, where
1
[0.1,0.9]w ∈
and
21
1ww=−
.
3 EXPERIMENTS
3.1 Data Sets and Experimental Setup
The gene expression data sets used in this article are
summarized in Table 1. They included binary-
classes and multi-classes data sets.
Table 1: The summary of gene expression data sets.
Data Sets
Number of
Samples
Number of
Genes
Number of
Classes
Leukemia 72 7,129 2
Colon 62 2,000 2
SRBCT 83 2,308 4
Note:
SRBCT = small round blue cell tumors.
DB = database.
DB Leukemia: http://www.broad.mit.edu/cgi-bin/cancer/datasets.cgi
DB Colon: http://microarray.princeton.edu/oncology/affydata/
index.html
DB SRBCT: http://research.nhgri.nih.gov/microarray/Supplement
All experimental results reported in this article are
experimented in Rocks Linux version 4.2.1
(Cydonia) on the IBM xSeries 335 (cluster nodes)
that contains 13 compute-nodes. Each compute-node
has four high performances 3.0GHz Intel Xeon
CPUs with 512MB of memories. Thus, the total
number of CPUs for the 13 compute-nodes is 52. In
order to make sure the running time of every run
using the same capacity of CPUs usage, each run has
been independently experimented on only one CPU.
This situation is important because the comparison
of running times between EPSO and BPSO is
conducted for evaluation of their performances.
Table 2: Parameter settings for EPSO and BPSO.
Parameters Values
The number of particles 100
The number of iterations (generations) 500
1
w 0.8
2
w 0.2
1
c 2
2
c 2
Experimental results that produced by EPSO are
compared with an experimental method (BPSO) and
other previous PSO-based methods for objective
comparisons (Shen et al., 2008; Chuang et al., 2008,
Li et al., 2008). SVM is used to measure LOOCV
accuracy on gene subsets that produced by EPSO
and BPSO. In order to avoid selection bias, the
implementation of LOOCV is in exactly the same
way as did by Chuang et al. (2008) where the only
one cross-validation cycle (outer loop), namely
LOOCV is used, not two nested ones. Several
experiments are independently conducted 10 times
on each data set using EPSO and BPSO. Next, an
average result of the 10 independent runs is
obtained. Two criteria following their importance
are considered to evaluate the performances of
EPSO and BPSO: LOOCV accuracy and the number
of selected genes. Additionally, running times are also
measured for the comparison between EPSO and
BPSO. High accuracy and the small number of
selected genes are needed to obtain an excellent
performance. Table 2 contains parameter values for
EPSO and BPSO. These values are chosen based on
the results of preliminary runs.
ICAART 2010 - 2nd International Conference on Agents and Artificial Intelligence
86

Table 3: Experimental Results for each Run Using EPSO on Leukemia, Colon, and SRBCT Data Sets.
Run#
Leukemia Colon SRBCT
#Acc
(%)
#Selected
Genes
#Time
#Acc
(%)
#Selected
Genes
#Time
#Acc
(%)
#Selected
Genes
#Time
1 100 55 7.61 93.55 17 5.16 100 33 9.91
2 100 65 7.37 93.55 11 4.93 100 26 9.91
3 100 65 7.40 95.16 22 4.98 100 25 10.36
4
100 70 7.42 96.77 22 4.94 100 26 10.37
5
100 51 7.52 98.39 23 5.06 100 31 7.31
6 100 62 7.48 95.16 15 5.07 100 22 7.31
7 100 58 7.43 93.55 27 5.00 100 26 10.33
8 100 61 7.45 95.16 29 5.02
100 21 10.32
9 100 63 7.46 93.55 20 5.03 100 29 10.24
10 100 67 7.46 91.94 16 5.02 100 22 10.24
Average
± S.D.
100
± 0
61.70
± 5.72
7.46
± 0.67
94.68
± 1.87
20.20
± 5.55
5.02
± 0.07
100
± 0
26.10
± 3.96
9.63
± 1.24
N
ote: The result of the best subsets is shown in the shaded cells. It is selected based on the following priority criteria: 1) the highest classification
a
ccuracy; 2) the smallest number of selected genes; 3) the lowest running time. #Acc and S.D. denote the classification accuracy and the standard
d
eviation, respectively, whereas #Selected Genes and Run# represent the number of selected genes and a run number, respectively. #Time stands for
r
unning time.
Table 4: Comparative experimental results of EPSO and BPSO.
Data
Method
Evaluation
EPSO BPSO
Best #Ave S.D Best #Ave S.D
Leukemia
#Acc (%)
100 100 0 98.61 98.61 0
#Genes
51 61.70 5.72 3488 3528.75 26.83
#Time
7.52 7.46 0.67 261.34 261.41 0.18
Colon
#Acc (%)
98.39 94.68 1.87 90.32 88.55 0.92
#Genes
23 20.20 5.55 982 985.00 25.22
#Time
5.06 5.02 0.07 64.45 64.63 0.18
SRBCT
#Acc (%)
100 100 0 100 100 0
#Genes
21 26.10 3.96 1076 1098.33 12.46
#Time
10.32 9.63 1.24 136.81 136.87 0.06
N
ote: The best result of each data set is shown in the shaded cells. It is selected based on the following priority criteria:
1) the highest classification accuracy; 2) the smallest number of selected genes; 3) the lowest running time.
Table 5: A comparison between our method (EPSO) and previous PSO-based methods.
Data
Method
Evaluation
EPSO
IBPSO
(Chuang
et al., 2008)
PSOTS
(Shen et al., 2008)
PSOGA
(Li et al., 2008)
Leukemia
#Acc (%)
(100) 100 (98.61) (95.10)
#Genes (61.70) 1034 (7) (21)
Colon
#Acc (%)
(94.68)
-
(93.55) (88.7)
#Genes
(20.20)
- (8) (16.8)
SRBCT
#Acc (%)
(100)
100 - -
#Genes
(26.10)
431 - -
Note: The result of the best subsets is shown in the shaded cells. It is selected based on the following priority criteria: 1) the highest classification
accuracy; 2) the smallest number of selected genes. ‘-‘ means that a result is not reported in the previous related work. A result in ‘( )’ denotes an
average result.
IBPSO = An improved binary PSO.
PSOGA = A hybrid of PSO and GA.
PSOTS = A hybrid of PSO and tabu search.
GPSO = Geometric PSO.
SELECTING GENES FROM GENE EXPRESSION DATA BY USING AN ENHANCEMENT OF BINARY PARTICLE
SWARM OPTIMIZATION FOR CANCER CLASSIFICATION
87
3.2 Experimental Results
Based on the standard deviation of classification
accuracy in Table 3, results that produced by EPSO
were consistent on all data sets. Interestingly, all
runs have achieved 100% LOOCV accuracy with
less than 71 selected genes on the Leukemia and
SRBCT the data sets. Moreover, over 91%
classification accuracies have been obtained on the
Colon data set. This means that EPSO has efficiently
selected and produced a near-optimal gene subset
from high-dimensional data (gene expression data).
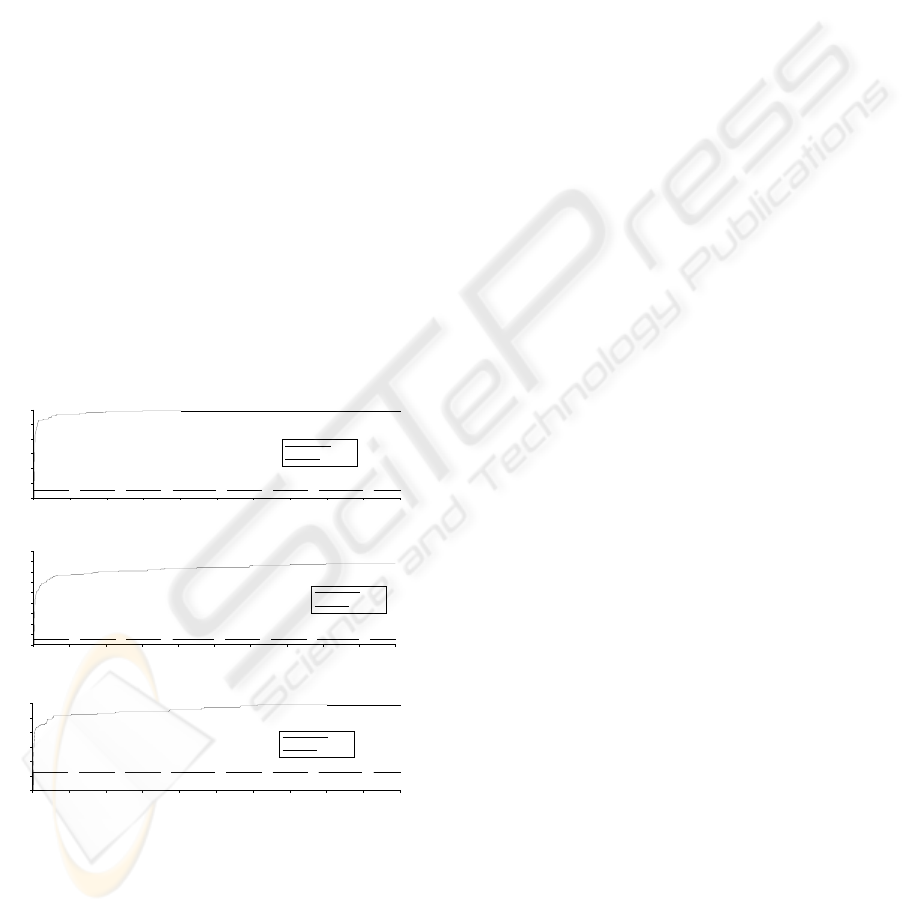
Figure 2 shows that the averages of fitness
values of EPSO increase dramatically after a few
generations on all the data sets. A high fitness value
is obtained by a combination between a high
classification rate and a small number (subset) of
selected genes. The condition of the proposed
particles’ speed that should always be positive real
numbers started in the initialization method, the new
rule for updating particle’s positions, and the
modified sigmoid function provoke the early
convergence of EPSO. In contrast, the averages of
fitness values of BPSO was no improvement until
the last generation due to
( ( ) 0) ( ( ) 1) 0.5.
dd
ii
Px t Px t== ==
Leukaemia Data Set
0.88
0.9
0.92
0.94
0.96
0.98
1
0 50 100 150 200 250 300 350 400 450 500
Generation
Fitness
EPSO
BPSO
SRBCT Data Set
0.88
0.9
0.92
0.94
0.96
0.98
1
0 50 100 150 200 250 300 350 400 450 500
Generation
Fitness
EPSO
BPSO
Colon Data Set
0.8
0.82
0.84
0.86
0.88
0.9
0.92
0.94
0.96
0.98
0 50 100 150 200 250 300 350 400 450 500
Generation
Fitness
EPSO
BPSO
Figure 2: The relation between the average of fitness
values (10 runs on average) and the number of generations
for EPSO and BPSO.
According to the Table 4, overall, it is
worthwhile to mention that the classification
accuracy of EPSO are superior to BPSO in terms of
the best, average, and standard deviation results on
all the data sets.. Moreover, EPSO also produces a
smaller number of genes compared to BPSO. The
running times of EPSO are lower than BPSO in all
the data sets. EPSO can reduce its running times
because of the following reasons:
EPSO selects the smaller number of genes
compared to BPSO;
The computation of SVM is fast because it uses
the small number of features (genes) that
selected by EPSO for classification process;
EPSO only uses the speed of a particle for
comparing with
3
()
d
rt, whereas BPSO practices
all elements of a particle’s velocity vector for the
comparison.
For an objective comparison, we compare our
work with previous related works that used PSO-
based methods in their proposed methods (Shen et
al., 2008; Chuang et al., 2008; Li et al., 2008). It is
shown in Table 5. For all the data sets, the averages
of classification accuracies of our work were higher
than the previous works. Our work also have
resulted the smaller averages of the number of
selected genes on the data sets compared to the
previous works. The latest previous work also came
up with the similar LOOCV results (100%) to ours
on the Leukemia and SRBCT data sets, but they
used many genes (more than 400 genes) to obtain
the same results (Chuang et al., 2008). Moreover,
they could not have statistically meaningful
conclusions because their experimental results were
obtained by only one independent run on each data
set, and not based on average results. The average
results are important since their proposed method is
a stochastic approach. Additionally, in their
approach, the global best particles’ position is reset
to zero position when its fitness values do not
change after three successive iterations.
Theoretically, their approach is almost impossible to
result a near-optimal gene subset from high-
dimensional spaces (high-dimension data) because
the global best particles’ position should make a new
exploration and exploitation for searching the near-
optimal solution after its position reset to zero.
Overall, our work has outperformed the previous
related works in terms of LOOCV accuracy and the
number of selected genes.
According to Fig. 3 and Tables 3-5, EPSO is
reliable for gene selection since it has produced the
near-optimal solution from gene expression data.
This is due to the proposed particles’ speed, the
introduced rule, and the modified sigmoid function
increase the probability
(1)0
d
i
xt+=
ICAART 2010 - 2nd International Conference on Agents and Artificial Intelligence
88

(( ( 1) 0))
d
i
Px t+=
. This high probability causes the
selection of a small number of informative genes
and finally produces a near-optimal subset (a small
subset of informative genes with high classification
accuracy) for cancer classification. The particles’
speed is introduced to provide the rate at which a
particle changes its position, whereas the rule is
proposed to update particle’s positions. The sigmoid
function is modified for increasing the probability of
bits in particle’s positions to be zero.
4 CONCLUSIONS
In this paper, EPSO has been proposed for gene
selection on three gene expression data sets. Overall,
based on the experimental results, the performance
of EPSO was superior to BPSO and PSO-based
methods that proposed by previous related works in
terms of classification accuracy and the number of
selected genes. EPSO was excellent because the
probability
(1)0
d
i
xt+=
has been increased by the
proposed particles’ speed, the introduced rule, and
the modified sigmoid function. The particles’ speed,
the introduced rule, and the modified function have
been proposed in order to yield a near-optimal
subset of genes for better cancer classification.
EPSO also obtains lower running times because it
selects the small number of genes compared to
BPSO. For future works, a modified representation
of particle’s positions in PSO will be proposed to
reduce the number of genes subsets in solution
spaces.
REFERENCES
Chuang, L. Y., Chang, H. W., Tu, C. J. and Yang, C. H.
(2008). Improved Binary PSO for Feature Selection
Using Gene Expression Data.
Computational Biology
and Chemistry
, 32, 29-38.
doi:10.1016/j.compbiolchem.2007.09.005
Kennedy, J. and Eberhart, R. 1995. Particle swarm
optimization. In
Proceeding of the 1995 IEEE
International Conference on Neural Networks
, 4,
1942-1948. IEEE Press. Retrieved from: IEEE Xplore
Digital Library.
Kennedy, J. and Eberhart, R. 1997. A discrete binary
version of the particle swarm algorithm. In
Proceeding
of the 1997 IEEE International. Conference on
Systems, Man, and Cybernetics
, 5, 4104-4108. IEEE
Press. Retrieved from: IEEE Xplore Digital Library.
Knudsen, S. (2002).
A Biologist’s Guide to Analysis of
DNA Microarray Data
(1
st
ed.). New York: John
Wiley & Sons.
Li, S., Wu, X. and Tan, M. (2008). Gene Selection Using
Hybrid Particle Swarm Optimization and Genetic
Algorithm.
Soft Computing, 12, 1039-1048.
doi:10.1007/s00500-007-0272-x
Mohamad, M. S., Omatu, S., Yoshioka, M. and Deris, S.
(2009). A Cyclic Hybrid Method to Select a Smaller
Subset of Informative Genes for Cancer Classification.
International Journal of Innovative Computing,
Information and Control
, 5(8), 2189-2202.
Shen, Q., Shi, W. M. and Kong, W. Hybrid Particle
Swarm Optimization and Tabu Search Approach for
Selecting Genes for Tumor Classification Using Gene
Expression Data.
Computational Biology and
Chemistry
, 32, 53-60. doi:10.1016/j.compbiolchem.
2007.10.001
SELECTING GENES FROM GENE EXPRESSION DATA BY USING AN ENHANCEMENT OF BINARY PARTICLE
SWARM OPTIMIZATION FOR CANCER CLASSIFICATION
89
