
INTELLIGENT CLINICAL DECISION SUPPORT SYSTEMS
Alexandru G. Floares
SAIA - Solutions of Artificial Intelligence Applications, Str. Vlahuta, Bloc Lama C/45, Cluj-Napoca, Romania
IOCN - Cancer Institute Cluj-Napoca, Artificial Intelligence Department, Str. Republicii, Nr. 34-36, Cluj-Napoca, Romania
Keywords:
Clinical decision support systems, Data mining, Artificial intelligence, Chronic hepatitis, Prostate cancer,
Biopsy.
Abstract:
Clinical Decision Support Systems (CDSS) have the potential to replace painful, invasive, and costly proce-
dures, to optimize medical decisions, improve medical care, and reduce costs. An even better strategy is to
make use of a knowledge discovery in data approach, with the aid of artificial intelligence tools. This results
in transforming conventional CDSS in Intelligent Clinical Decision Support (i-CDSS). Evolving i-CDSS give
to the conventional CDSS the capability of self-modifying their rules set, through supervised learning from
patients data. Intelligent and evolving CDSS represent a strong foundation for evidence-based medicine. We
proposed a methodology of building i-CDSS and related concepts. These are illustrated with some of our
results in liver diseases and prostate cancer, some of them showing the best published performance.
1 INTRODUCTION AND
BACKGROUND
The use of information technology for replacing
painful, invasive, and/or costly procedures, for opti-
mizing various medical decisions, or for improving
medical care and reducing the costs, represent major
goals of Medical Informatics. Implementing Clini-
cal Decision Support Systems (CDSS) (Berner, 2007)
on a large scale is a major step toward these goals.
Using a knowledge discovery in data approach with
artificial intelligence tools, one can build Intelligent
Clinical Decision Support Systems (i-CDSS) instead
of conventional CDSS.
We performed a set of investigations on con-
structing i-CDSS for several liver diseases, prostate
and thyroid cancer, and chromosomal disorders (e.g.,
Down syndrome) during pregnancy. The high perfor-
mance of the liver and prostate i-CDSS determined
us to consider some of these methods and concepts
mature and general enough to be presented, but still
under development. Here, we present a methodology
of building i-CDSS and the related concepts. These
are illustrated with some of our results in liver and
prostate diseases, showing the best published perfor-
mances out to date, to our knowledge.
Chronic Hepatitis B and C are major diseases of
mankind and a serious global public health problem.
The persons with these chronic diseases are at high
risk of death from cirrhosis and liver cancer. Liver
biopsy is the gold standard for grading the severity
of disease, and staging the degree of fibrosis, and the
grade of necroinflammation. The most used scoring
systems are:
1. METAVIR A (A stands for activity) or Ishak NI
(NI stands for necroinflammatory) for necroin-
flammatory grade
2. METAVIR F or Ishak F for the fibrosis stage (F
stands for fibrosis).
By assigning scores for severity, grading, and staging
of hepatitis, they are very important for patient man-
agement.
Liver biopsy is invasive, painful, and relatively
costly; complications severe enough to require hos-
pitalization can occur in about 4% of patients (Lin-
dor, 1996). In a review of over 68,000 patients re-
covering from liver biopsy, 96% experienced adverse
symptoms during the first 24 hours of recovery. Hem-
orrhage was the most common symptom, but infec-
tions also occurred. Side effects of the biopsies in-
cluded pain, tenderness, internal bleeding, pneumoth-
orax, and rarely, death (Tobkes and Nord, 1995)
There are two main non-invasive diagnosis tech-
niques of interest (Shaheen et al., 2007). FibroScan is
282
G. Floares A. (2010).
INTELLIGENT CLINICAL DECISION SUPPORT SYSTEMS.
In Proceedings of the Third International Conference on Health Informatics, pages 282-287
DOI: 10.5220/0002740802820287
Copyright
c
SciTePress

a type of ultrasound machine that uses transient elas-
tography to measure liver stiffness. The device re-
ports a value that is measured in kilopascals (kPa). Fi-
broTest for assessing fibrosis, and ActiTest for assess-
ing necroinflammatory activity are available through
BioPredictive (www.biopredictive.com). These tests
use algorithms to combine the results of serum tests
of beta 2-macroglobulin, haptoglobulin, apolipopro-
tien A1, total bilirubin, gamma glutamyltranspepti-
dase (GGT), and alanine aminotransferase (ALT).
The results of these diagnosis techniques are not
directly interpretable by a pathologist, but can be ex-
trapolated to a fibrosis and necroinflammation score .
FibroTest and FibroScan have reasonably good utility
for the identification of cirrhosis, but lesser accuracy
for earlier stages. It is considered that refinements are
necessary before these tests can replace liver biopsy
(Shaheen et al., 2007).
We used a knowledge discovery in data, based on
artificial intelligence, to investigatethe possibilities of
accuracy improvements and of expressing the results
in the pathologist scoring systems.
In our prostate cancer studies, one goal was to in-
vestigate the possibility of developing a non-invasive
diagnosis i-CDSS, based mainly on the concentra-
tions of a set of 8 angiogenic molecules in serum.
A detailed description of these data can be found in
(Balacescu et al., 2008). For the purpose of this pa-
per, which is to outline the methodology and the con-
cepts related to evolving intelligent CDSS, unnec-
essary molecular biology, medical and data mining
technicalities were eliminated.
To our knowledge, this is the first study, inte-
grating angiogenic molecules, clinical and laboratory
data, to develop intelligent systems capable to predict
diagnosis like prostate cancer or benignant diseases,
which are usually based on the prostate biopsy. The
preliminary results are very encouraging, with an ac-
curacy ranging from 97.8% to 100% (manuscript in
preparation).
2 DEVELOPING INTELLIGENT
CLINICAL DECISION
SUPPORT: A METHODOLOGY
AND RELATED CONCEPTS
In essence, we extracted and integrated information
from various non-invasive data sources, e.g. imag-
ing, clinical, routine laboratory or molecular data, and
build i-CDSS capable to predict various results of the
biopsy, e.g., liver fibrosis or prostate cancer diagnosis,
with an acceptable accuracy. The meaning of accept-
able accuracy depends on the specific medical context
and is a matter of consensus. Probably, in this context
the prediction accuracy should be at least 80%.
Because the results of the liver, prostate, or other
organs biopsy, are used in many important medical
decisions, in the management of the related diseases,
we investigated the possibility of developing other i-
CDSS, starting from these. For example, important
treatment decision are partially based on biopsy.
Chronic hepatitis B and C are treated with drugs
called Interferon or Lamivudine, which can help some
patients. This treatment decision is based on several
patients selection criteria. For example, the criteria
for selecting the patients with chronic hepatitis C who
will benefit from Interferon treatment, are:
1. Chronic infection with hepatitis C virus (HCV):
antibodies against HCV (anti-HCV) are present
for at least 3 months.
(a) the hepatitis B surface antigen (HBsAg) is
present for at least 6 months, or
(b) the hepatitis B e antigen (HBeAg) is present for
at least 10 weeks.
2. The cytolytic syndrome: the transaminases level
is increased or normal.
3. Pathology (biopsy): the Ishak NI ≥ 4 and Ishak F
≤ 3.
4. The virus is replicating: the transaminases level
is increased or normal, and anti-HCV are present,
and RNA-HCV ≥ 10
5
copies/milliliter.
For hepatitis B there is a similar set of selection
rules.
Analyzing these treatment decisions, one can
identify two problems:
1. Invasiveness: the patients selection criteria in-
clude fibrosis and necroinflammation assessed by
liver biopsy, an invasive medical procedure.
2. Cost of the wrong decisions: patients selec-
tion errors are very costly, because Interferon or
Lamivudine therapy costs thousands of dollars.
In the methodological context proposed in this
paper, developing solutions to these problems is
straightforward. The aforementioned conditions are
easy to implement in an interactivecomputer program
and biopsy could be replaced by the non-invasive i-
Biopsy. Developing intelligent CDSS, based on non-
invasive medical investigations, and optimized selec-
tion criteria, could be of great benefit to the patients
and could also save money.
More precisely, one should investigate if it is pos-
sible:
INTELLIGENT CLINICAL DECISION SUPPORT SYSTEMS
283

1. To build i-CDSS capable to predict the biopsy
results—fibrosis stage and necroinflammation
grade—with an accuracy of at least 80%.
2. To integrate the i-CDSS predicting the biopsy re-
sults with the other selection criteria in an i-CDSS
for Interferon treatment.
3. To make the Interferon treatment i-CDSS an
evolving one, capable to optimize the treatment
decisions by self-modifying through learning.
It is interesting to note that some of the compo-
nents of the treatment i-CDSS are fixed in time, while
other can evolve, by learning from data. In this exam-
ple, the i-Biopsy component is fixed, being an input-
output relationship, already discovered from patients
data, and used to predict the results of the biopsy,
without performing it. The component implementing
the treatment selection criteria could be either fixed or
evolving through learning. In the first case we have a
fixed i-CDSS, and in the second case an evolving one.
Evolving i-CDSS can minimize the costs due to
erroneous patients selection, and maximize the ben-
efit of the treatment. They can optimize the selec-
tion rule sets by finding the relevant selection criteria
and their proper cutoff values. For this, the outcomes
of the Interferon treatment must be clearly defined as
numerical or categorical attributes and registered in a
data base for each treated patient.
Then, intelligent agents are employed to learn the
prediction of the treatment outcomes. They must
be capable of expressing the extracted information
as rules, using non-invasive clinical, laboratory and
imaging attributes as inputs. Using feature selection
(see for example (Guyon et al., 2006)) one will find
the relevant patient selection criteria.
Thus, the i-CDSS started with the accepted pa-
tients selection criteria, but these are evolving. It is
worth to mention that the evolved selection criteria
could be different, from those initially proposed by
physicians, and usually better. However, they should
be always evaluated by the experts. In the supervised
learning process, intelligent agents also discover the
proper cutoff values of the relevant selection criteria.
Again, these are usually better than those proposed
by experts, but they should always be evaluated by
them. In our opinion, evolving through learning from
patients data is crucial for evidence based-medicine.
These i-CDSS are the result of a data mining pre-
dictive modeling strategy, which is now patent pend-
ing, consisting mainly in:
1. Extracting and integration information from var-
ious medical data sources, after a laborious pre-
processing:
(a) cleaning features and patients,
(b) various treating of missing data,
(c) ranking features,
(d) selecting features,
(e) balancing data.
2. Testing various classifiers or predictive modeling
algorithms.
3. Testing various ensemble methods for combining
classifiers.
For modeling, we tested the prediction accuracy of
various types of artificial intelligence agents:
1. Neural Networks of various types and architec-
tures,
2. Decision trees C5.0 and Classification and Re-
gression Trees
3. Support Vector Machines, with various kernels
4. Bayesian Networks
5. Genetic Programming based agents.
i-Biopsy is an intelligent system based on any algo-
rithm or combination of algorithms capable of learn-
ing from data. Of course, accuracy is very important
but physicians also prefer white-box algorithms and
transparent decisions.
We have chosen C5.0 decision trees, the last ver-
sion of the C4.5 algorithm (Quinlan, 1993), with
10-fold cross-validation. As ensemble method, we
used Freund and Schapire’s boosting (Freund and
Schapire, 1997) for improving the predictive power of
C5.0 classifier learning systems. A set of C5.0 classi-
fiers is produced and combined by voting, and by ad-
justing the weights of training cases. We suggest that
boosting should always be tried when peak predictive
accuracy is required, especially when unboosted clas-
sifiers are already quite accurate .
Genetic programming was another important
choice, giving accurate and transparent i-CDSS in the
form of mathematical models of the input-output rela-
tionship (manuscript in preparation). Transparency of
the i-CDSS is affected by boosting and is less useful
when the number of variables is large.
3 MAIN RESULTS
In what follows, some of the results illustrating the
practical and conceptual significance of i-Biopsy are
presented. The examples are i-CDSS selected from
gastroenterology and urology.
In one of our hepatological studies, we collected a
dataset of 700 chronic hepatitis C patients and about
135 inputs. One of the i-CDSS has liver fibrosis as the
HEALTHINF 2010 - International Conference on Health Informatics
284
predicted output expressed as Metavir F score, having
five classes: from Metavir F0 to Metavir F4. The ac-
curacy of the first experiments was about 60%. Pre-
processing increased the accuracy with 20% to 25%.
As we mentioned, we tested various algorithms and
settings, but C5.0 accuracy was one of the highest,
about 80% (see Table 1 and Table 2 for some exam-
ples).
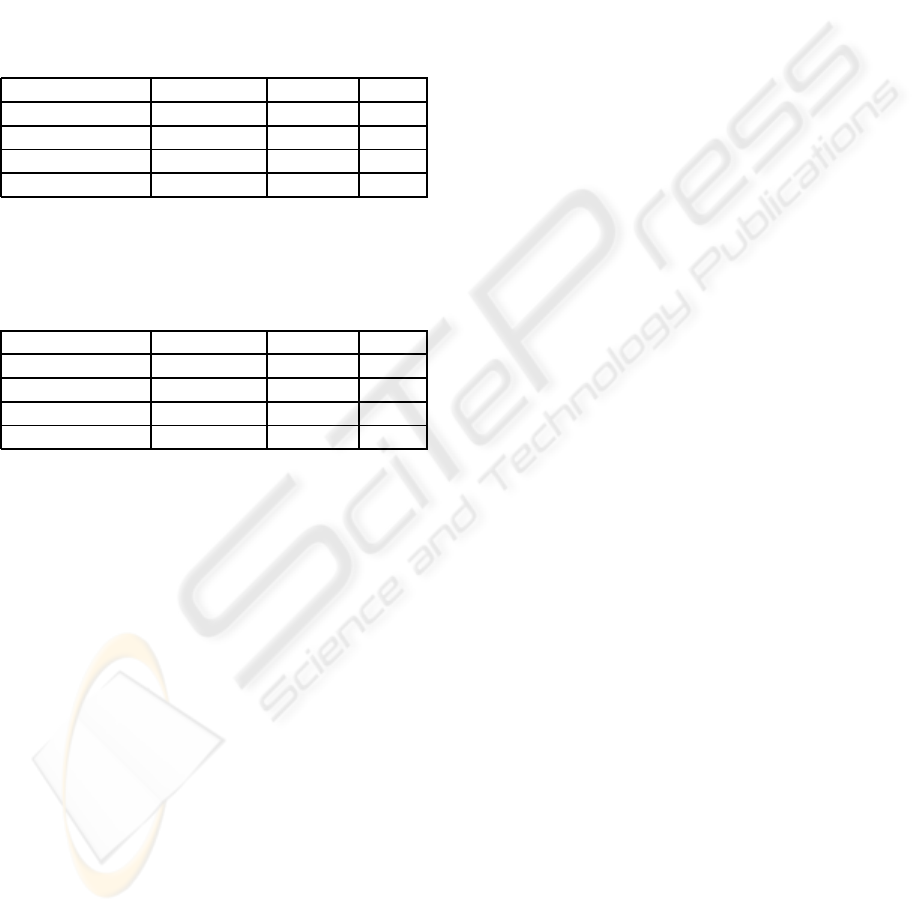
Table 1: Results of experiments for feature and algorithm
selection for METAVIR F0 prediction, formulated as a one-
versus-all classification (Logistic Regres stands for Logistic
Regression; AUC stands for Area Under the Curve).
Model Accuracy% Features AUC
C5 99.639 19 1
CART 95.307 17 0.755
Logistic Regres 93.863 25 0.918
Neural Net 92.78 25 0.648
Table 2: Results of experiments for feature and algorithm
selection for METAVIR F4 prediction, formulated as a one-
versus-all classification (Logistic Regres stands for Logistic
Regression; AUC stands for Area Under the Curve).
Model Accuracy% Features AUC
C5 95.668 11 0.896
CART 94.224 21 0.914
Logistic Regres 92.419 25 0.906
Neural Net 88.809 25 0.867
Parameter tuning and boosting increase the accu-
racy of some i-CDSS even to 100% (Floares et al.,
2008; Floares, 2009b).
We also developed liver i-Biopsy versions based
on Bayesian Networks and on Genetic Programming,
some of them as binary classifiers (work in progress).
After many experiments, we conclude that important
is to reach the highest robust accuracy, between 80%
and 100%, and the test for this is the external valida-
tion.
Because the results of the biopsy are central
to important medical decisions, in the management
of chronic hepatitis patients, it was relatively easy
to build i-CDSS for Interferon treatment (Floares,
2008), (Floares, 2009a). This was done just by adding
the aforementioned patients selection criteria to the
i-Biopsy. This non-invasive i-CDSS is of a special
kind; being able to evolve, by attempting to predict
the progressively accumulating outcomes of the Inter-
feron treatment, it will eventually identify the proper
patients selection criteria, and their cutoff values from
data (see section 2 for more details). Thus, the rules
set of this i-CDSS is evolving.
We tried to develop not only the technical aspects
of the intelligent CDSS, evolving through learning
from data, but also the related concepts.
i-Biopsy, one of the central concepts, is an intelli-
gent system (the prefix ”i-” coming from intelligent),
supporting important medical decisions, by being ca-
pable to predict, with an acceptable accuracy, the re-
sults usually givenby a pathologist, examining the tis-
sue samples from biopsies. Real biopsy is performed
on different organs, e.g., liver, prostate, etc., and the
pathologists expressed their findings as diagnoses or
scores of a largely accepted scoring system. While the
concept is general, individual systems must be spe-
cific (see below).
For example, let us shortly analyze liver (organ) i-
Biopsy (the intelligent counterpart of the real biopsy),
in chronic hepatitis C (disease), assessing liver fibro-
sis (diagnose), expressed by METAVIR F (pathologist
scoring system). This liver i-Biopsy takes as inputs
and integrate various routine, non-invasive, clinical,
imaging and lab data.
To distinguish between the scores of the real
biopsy and their counterparts predicted by i-Biopsy,
we proposed the general terms of i-scores. There are
many examples, like Gleason score in prostate cancer,
but we continue to focus on the gastroenterological
applications, where we have:
1. The liver i-Biopsy is the i-CDSS correspond-
ing to the real liver biopsy; the i-METAVIR F
scores are the values predicted by i-Biopsy for the
METAVIR-F fibrosis scores, designating exactly
the same pathological features.
2. The i-METAVIR F scores and the biopsy
METAVIR F scores could have different values
for the same patient, at the same moment, de-
pending for example on the prediction accuracy
or pathologist errors.
3. i-METAVIR F scores are obtained in a non-
invasive, painless, and riskless manner, as op-
posed to METAVIR-F scores, assessed by liver
biopsy.
For simplicity, we referred only to the METAVIR
F scores, but these considerations are general, and can
be easily extrapolated to other liver scores, like Ishak
F, METAVIR A, and Ishak NI. Moreover, these con-
ceptual clarifications apply to any situation in which
the following elements are present:
1. an anatomical structure, e.g., liver, or prostate, etc.
2. the invasive procedure, e.g., biopsy
3. a disease, e.g., chronic hepatitis C or B, prostate
cancer or benign prostatic hyperplasia
4. a set of pathological features or diagnoses to as-
sess, e.g., fibrosis, necroinflammation, etc.
INTELLIGENT CLINICAL DECISION SUPPORT SYSTEMS
285

5. a set of classes for the pathological findings, e.g.
Gleason or METAVIR scores, pathological diag-
noses, etc.
The second element (biopsy) could be replaced,
at least in a considerable percentage of cases, by the
i-Biopsy, and the fifth element by the i-scores.
We have built the following i-CDSS which can be
used for Interferon treatment decision support:
1. Module for liver fibrosis prediction,
(a) according to METAVIR F scoring system, with
and without liver stiffness (FibroScan),
(b) according to Ishak F scoring system, with and
without liver stiffness (FibroScan).
2. Module for the grade of necroinflammation (ac-
tivity) prediction, according to Ishak NI scoring
systems.
Also, we developed some prostate i-Biopsy sys-
tems, as non-invasive i-CDSS counterparts for some
prostate biopsy results. For example, i-Gleason score
is the i-Biopsy predicted Gleason score (work in
progress), and is also central to many important med-
ical decisions in prostate cancer. The three classes
classifiers, distinguishing between normal, benignant
and malignant are more interesting for the fundamen-
tal research. The binary classifiers, especially those
distinguishing between malignant and benignant, are
clinically oriented. The preliminary results are very
encouraging, with accuracy ranging from 97.8% to
100%.
4 DISCUSSION
A short digression about the meaning of the diagno-
sis accuracy, of the i-CDSS in general and i-Biopsy in
particular, seems necessary, because it confused many
physicians, especially when reporting very high val-
ues like 100%. Many physicians believe that 100%
accuracy is not possible in medicine. The mean-
ings will be made clear trough examples. Typically,
an invasive liver (or prostate, etc.) biopsy is per-
formed to the patient, and a pathologist analyzes the
tissue samples assessing fibrosis, necroinflammation,
etc., and expressed the results as scores or patholog-
ical diagnosis. The pathologist may have access to
other patient medical data, but usually these are not
necessary for him or her to formulate the patholog-
ical diagnosis. Moreover, in some studies it is re-
quired that the pathologist knows nothing about the
patient. His or her diagnosis can be more or less cor-
rect or even wrong, for many reasons not discussed
here. We have proposed i-CDSS predicting the fibro-
sis scores resulted from liver biopsy, or the prostate
cancer diagnosis resulted from prostate biopsy, with
accuracy reaching 90% - 100%. For the i-CDSS, sev-
eral clinical, imaging, and lab data of the patient are
essential, because they were somehow incorporated
in the system. They were used like input features
to train the system, and they are required for a new,
unseen patient, because i-Biopsy is a relationship be-
tween these inputs and the fibrosis, necroinflamma-
tion scores, or diagnosis as outputs. The category of i-
CDDSs discussed here do not deal directly with diag-
nosis correctness, but with diagnosis prediction accu-
racy. Without going into details, this is due in part to
the supervised nature of the learning methods used to
build them. The intelligent agents learned to predict
the results of the biopsy given by the pathologist, and
the pathologist diagnosis could be more or less cor-
rect. For example, let us suppose that the pathologist
diagnosis is wrong. The i-Biopsy could still be 100%
accurate in predicting this wrong diagnosis, but this
is rarely the case. In other words, the i-Biopsy will
predict, in a non-invasive and painless way, and with-
out the risks of the biopsy, a diagnosis which could be
even 100% identical with the pathologist diagnosis,
if the biopsy is performed. While the accuracy and
the correctness of the diagnosis are related in a sub-
tle way, they are different matters. i-Biopsy will use
the information content of several non-invasiveinves-
tigations, to predict the pathologist diagnosis, without
performing the biopsy. The correctness of the diagno-
sis is a different matter, but typically a good accuracy
correlates well with a correct diagnoses. The accu-
racy of the diagnosis, as well as other performance
measures like the area under the receiver operating
characteristic (AUROC), for a binary classifier sys-
tem (Fawcett, 2004), are useful for intelligent sys-
tems comparison. To our knowledge, the proposed
liver i-Biopsy system outperformed the most popu-
lar and accurate system, FibroTest and ActiTest (Sha-
heen et al., 2007) commercialized by BioPredictive
company, and FibroScan. The liver i-Biopsy is a
multi-classes classifier, expressing the results in the
pathologist’s scoring systems, e.g., five classes for
METAVIR F and seven classes for Ishak F. Multi-
classes classifiers are more difficult to develop than
binary classifiers, with outputs not directly related to
the fibrosis scores. We also build binary classifiers as
decision trees with similar accuracy and mathemati-
cal models (work in progress). Despite the fact that
AUROC is only for binary classifiers, loosely speak-
ing a 100% accuracy n classes classifier is equivalent
with n binary classifiers with AUROC = 1 (maximal).
BioPredictive company analyzed a total of 30 studies
HEALTHINF 2010 - International Conference on Health Informatics
286

(Poynard et al., 2007) which pooled 6,378 subjects
with both FibroTest and biopsy (3,501 chronic hep-
atitis C). The mean standardized AUROC was 0.85
(0.82-0.87). The robustness of these results is clearly
demonstrated by this cross-validation, while i-Biopsy
results need to be cross-validated. The fact that i-
Biopsy , in its actual setting, relies on routine ul-
trasound features is both a strong point and a weak
one, because of the subjectiveness in ultrasound im-
ages interpretation. It is worth to note that in certain
circumstances the result of the liver i-Biopsy could
be superior to that of real biopsy. When building
the i-CDSS, the results of the potentially erroneous
biopsies, which are not fulfilling some technical re-
quirements, were eliminated from the data set. Thus,
the i-Biopsy predicted results correspond only to the
results of the correctly performed biopsies, while a
number of of the real biopsy results are wrong, be-
cause they were not correctly performed. Due to the
invasive and unpleasant nature of the biopsy, is very
improbable that a patient will accept a technically in-
correct biopsy to be repeated. Unlike real biopsy,
i-Biopsy can be used to evaluate fibrosis evolution,
which is of interest in various biomedical and pharma-
ceutical studies, because, being non-invasive,painless
and without any risk, can be repeated as many time as
needed.
Also, in the early stages of liver diseases, often the
symptoms are not really harmful for the patient, but
the treatment is more effective than in more advanced
fibrosis stages. The physician will hesitate to indicate
an invasive, painful and risky liver biopsy, and the pa-
tients are not as worried about their disease as they are
about the pain of the biopsy. However, i-Biopsy can
be performed and an early start of the treatment could
be much more effective.
ACKNOWLEDGEMENTS
We thank to the following medical teams: Dr. Mon-
ica Lupsor, Dr. T. Suteu and Prof. Dr. R. Badea, from
Medical Imaging Department, Dr. H. Stefanescu and
Dr. Z. Sparchez, from Hepatology Department, Dr.
A. Serban from Pathology Department, Dr. N. Crisan,
Dr. B. Feciche and Prof. Dr. I. Coman, from Urology
Department, University of Medicine and Pharmacy
Cluj-Napoca, Romania, Dr. Carmen Floares, Dr. O.
Balcescu, Dr. Ioana Neagoe Dr. Loredana Balacescu,
Dr. Oana Tudoran, and Prof. Dr. A. Irimie, from Can-
cer Institute Cluj-Napoca, Romania. We also thank
to the computer science team: Dr. F. Manolache, E.
Suica, and T. Popa.
REFERENCES
Balacescu, O., Neagoe, I., Balacescu, L., Crisan, N., Feci-
che, B., Tudoran, O., Coman, I., and Irimie, A. (2008).
Angiogenesis serum protein quantifcation for prostate
pathology. Curr Urol, (2):181–187.
Berner, E., editor (2007). Clinical decision support systems.
Springer: New York, NY.
Fawcett, T. (2004). Roc graphs: Notes and practical con-
siderations for researchers. technical report. Technical
report, Palo Alto, USA: HP Laboratories.
Floares, A. G. (2008). Intelligent systems for interferon
treatment decision support in chronic hepatitis c based
on i-biopsy. In IDAMAP - Intelligent Data Analysis in
Biomedicine and Pharmacology, Washington DC.
Floares, A. G. (2009a). Intelligent clinical decision supports
for interferon treatment in chronic hepatitis c and b
based on i-biopsy. In International Joint Conference
on Neural Networks, 2009, Atlanta, Georgia, USA.
Floares, A. G. (2009b). Liver i-Biopsy and the Correspond-
ing Intelligent Fibrosis Scoring Systems: i-Metavir F
and i-Ishak F, pages 253–264. Lecture Notes in Com-
puter Science. Springer Berlin / Heidelberg.
Floares, A. G., Lupsor, M., Stefanescu, H., Sparchez, Z.,
Serban, A., Suteu, T., and Badea, R. (2008). Toward
intelligent virtual biopsy: Using artificial intelligence
to predict fibrosis stage in chronic hepatitis c patients
without biopsy. Journal of Hepatology, 48(2).
Freund, Y. and Schapire, R. E. (1997). A decision theoretic
generalization of on line learning and an application to
boosting. Journal of Computer and System Sciences,
55(1):119–139.
Guyon, I., Gunn, S., Nikravesh, M., and Zadeh, L. (2006).
Feature Extraction: Foundations and Applications.
Studies in Fuzziness and Soft Computing. Springer.
Lindor, A. (1996). The role of ultrasonography and
automatic-needle biopsy in outpatient percutaneous
liver biopsy. Hepatology, 23:1079–1083.
Poynard, T., Morra, R., Halfon, P., Castera, L., Ratziu, V.,
Imbert-Bismut, F., Naveau, S., Thabut, D., Lebrec, D.,
Zoulim, F., Bourliere, M., Cacoub, P., Messous, D.,
Muntenau, M., and de Ledinghen, V. (2007). Meta-
analyses of fibrotest diagnostic value in chronic liver
disease. BMC Gastroenterology, 7(40).
Quinlan, J. (1993). C4.5 : Programs for Machine Learning.
Morgan Kaufmann.
Shaheen, A., Wan, A., and Myers, R. (2007). Fibrotest and
fibroscan for the prediction of hepatitis c-related fibro-
sis: a systematic review of diagnostic test accuracy.
Am J Gastroenterol, 102(11):2589–2600.
Tobkes, A. and Nord, H. J. (1995). Liver biopsy: Review of
methodology and complications. Digestive Disorders,
13:267–274.
INTELLIGENT CLINICAL DECISION SUPPORT SYSTEMS
287
