
MULTISPECTRAL TEXTURE ANALYSIS USING LOCAL BINARY
PATTERN ON TOTALLY ORDERED VECTORIAL SPACES
Vincent Barra
LIMOS, UMR 6158, Campus des C
´
ezeaux, 63173 Aubi
`
ere, France
Keywords:
Local binary pattern, Multispectral image, Segmentation, Texture, Total orderings.
Abstract:
Texture is an important feature when considering image segmentation. Since more and more image segmen-
tation problems involve multi- and hyperspectral data, including color images, it becomes necessary to define
multispectral texture features. In this article, we propose LMBP, an extension of the classical Local Binary
Pattern (LBP) operator to the case of multispectral images. The LMBP operator is based on the definition of
total orderings in the image space and on an extension of the standard univariate LBP. It allows the computa-
tion of both a multispectral texture structure coefficient and a multispectral contrast parameter for each spatial
location, that serve as an input to an unsupervised clustering algorithm. Results are demonstrated in the case of
the segmentation of brain tissues from multispectral MR images, and compared to other multispectral texture
features.
1 INTRODUCTION
Texture analysis plays an important role in several ap-
plications, from remote sensing to medical image pro-
cessing, industrial applications or document process-
ing. Four major issues are expected in texture anal-
ysis: feature extraction, texture discrimination, tex-
ture classification and shape from texture. To achieve
these analysis, several methods are available, using
statistical (autocorrelation, co-occurrence), geomet-
rical (structural techniques), model-based (MRF or
fractal) or signal processing based approaches (spa-
tial,Fourier, Gabor or wavelet filtering) (see for exam-
ple (Tuceryan and Jain, 1998) for a review). Among
all these methods, the Local Binary Pattern (LBP) op-
erator offers an efficient way of analyzing textures
(Ojala et al., 2002). It relies on a simple but efficient
theoretical framework, and combines both structural
and statistical properties.
In the prementioned potential applications, color and
even multispectral data is more and more available
and the extension of texture analysis methods be-
comes natural. We propose in this article to extend
the LBP operator to the case of multispectral images.
Contrary to other works (Lucieer et al., 2005; Song
et al., 2006), we do not apply the univariate LBP on
scalar values derived from the multispectral dataset,
but directly propose a multispectral LBP operator,
based on a total ordering computed either in the im-
ages space, or in a derived vectorial space.
The paper is organized as follows: Section 2 first
recall the original univariate LBP operator, and re-
call some basic definitions on orderings in a vectorial
space. It then introduces the LMBP - Local Multi-
spectral Binary Pattern, that uses both of these no-
tions. Section 3 presents and analyzes some prelim-
inary results of the operator on data stemming from
multispectral Magnetic Resonance Images.
2 LOCAL MULTISPECTRAL
BINARY PATTERN
2.1 Local Binary Pattern
Ojala et al. (Ojala et al., 2002) described the texture
T as the joint distribution of the gray levels of P + 1
image pixels: T = t(g
c
,g
0
···g
p−1
), where g
c
is the
gray level value of the center pixel, surrounded by P
equally spaces pixels of gray levels g
p
, located on a
circle of radius R. Gray values g
p
were interpolated
if neighbors didn’t fit on the pixel grid. They then
defined the Local Binary Pattern (LBP), a grayscale
invariant and rotation invariant operator:
LBP
riu2
P,R
=
P−1
∑
i=0
σ(g
p
− g
c
) if U(LBP
P,R
) ≤ 2
P + 1 otherwise
where
37
Barra V. (2010).
MULTISPECTRAL TEXTURE ANALYSIS USING LOCAL BINARY PATTERN ON TOTALLY ORDERED VECTORIAL SPACES.
In Proceedings of the International Conference on Computer Vision Theory and Applications, pages 37-43
DOI: 10.5220/0002828700370043
Copyright
c
SciTePress

U(LBP
P,R
) = |σ(g
P−1
− g
c
) − σ(g
0
− g
c
)|
+
P−1
∑
i=1
|σ(g
i
− g
c
) − σ(g
i−1
− g
c
)|
and σ(.) is the sign function. The uniformity function
U(LBP
P,R
) corresponds to the number of spatial tran-
sitions in the neighborhood: the larger it is, the more
likely a spatial transition occurs in the local pattern.
If LBP
riu2
P,R
captures the spatial structure of the texture,
it does not handle the strength of the pattern. To do
so, a contrast measure was defined as
C
P,R
=
1
P
P−1
∑
i=0
(g
i
− ¯g)
2
where ¯g =
1
P
P−1
∑
i=0
g
i
and textures may then be characterized by the joint
distribution of LBP
riu2
P,R
and C
P,R
.
Several extensions have been proposed to these fea-
tures (e.g. multi-resolution LBP (M
¨
aenp
¨
a
¨
a and
Pietik
¨
ainen, 2003; Ojala et al., 2002), center-
symmetric local binary pattern (Heikkil
¨
a et al.,
2009)), and numerous applications have been
adressed using these techniques (e.g. face recogni-
tion (Ahonen et al., 2006), segmentation of remote-
sensing images (Wang and Wang, 2006), visual in-
spection (Paclik et al., 2002) or classification of out-
door images (Garc
´
ıa and Puig, 2007)).
The LBP operator relies on the sign function σ(.),
and then on an ordering relation on the gray level
space. Since there is no natural ordering for vector
spaces, such as those produced by multispectral imag-
ing, the extension of LBP to multispectral data is not
straightforward. Some authors already defined mul-
tispectral LBP by combining intra-plane and inter-
plane LBP relations (Lucieer et al., 2005), or by con-
sidering the univariate LBP on vector norms (Song
et al., 2006), but to our knowledge no LBP opera-
tor has directly be defined on vectorial data. We thus
propose in the following to define the LMBP - Lo-
cal Multispectral Binary Pattern - operator, based on
LBP and on total orderings on R
n
. We first recall ba-
sic definitions on orders and then introduce the LMBP
operator.
2.2 Total Orderings in R
n
We first recall some basic definitions.
Definition 1. Let ≤
P
be a binary relation on a set P.
≤
P
is a pre-order if it is:
• reflexive: (∀x ∈ P) x ≤
P
x
• transitive: for all x,y,z ∈ P, if x ≤
P
y and y ≤
P
z
then x ≤
P
z
Definition 2. Let ≤
P
be a binary relation on a set
P. ≤
P
is a partial order if it is a pre-order and for all
x,y ∈ P, if x ≤
P
y and y ≤
P
x then x = y (antisymmetry)
Definition 3. Let ≤
P
be a partial order on a set P. ≤
P
is a total order if and only if for all x,y ∈ P, x ≤
P
y or
y ≤
P
x
If it is straightforward to define orders for scalar
values, the definition of partial -or total- orders for
vector valued data is not so easy: if data stems from
RGB images (P = R
3
), each channel being coded on
8 bits, each pixel can have one of the 2
24
possible
vectorial values, hence defining 2
24
! possible total or-
derings.
One has then to find another way to introduce order
in R
n
. Barnett (Barnett, 1976) defined four ways to
order vectors: the marginal approach, the partial ap-
proach, the conditional order and the dimension re-
duction. This last technique is an usual way to pro-
ceed, (Goutsias et al., 1995), and consists either in
defining an order using a distance in R
n
of each vec-
tor to a reference, or in projecting vectors into a vec-
torial space R
q
where an order can be defined. In this
latter case, the projection is defined by an applica-
tion h : R
n
→ R
q
, and (Chanussot and Lambert, 1998)
proved that :
• h defines an ordering relation ≤
n
in R
n
if and only
if h is injective (and h can be supposed to be bi-
jective if R
q
is restricted to h(R
n
))
• ≤
n
defines a total order in R
n
if and only if there
exist h : R
n
→ R
q
bijective defining ≤
n
on R
n
and
q=1
• ≤
n
defines a total order in R
n
if and only if ≤
n
defines a space filling curve of R
n
Total ordering may then be identically handled by the
definition of h, or by the construction of a space fill-
ing curve of R
n
. In the following, we propose as a
preliminary study to define ≤
n
using an appropriate
h.
2.3 The LMBP Operator
Figure 1 presents an overview of the algorithm. Each
step is detailed in the following subsections
2.3.1 Subspace Analysis
Since numerous information may be available from
the original dataset of R
p
, and since the p original
images may be dependent, it may be useful to con-
duct feature extraction before defining the vector or-
dering. Several techniques are available to extract
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
38
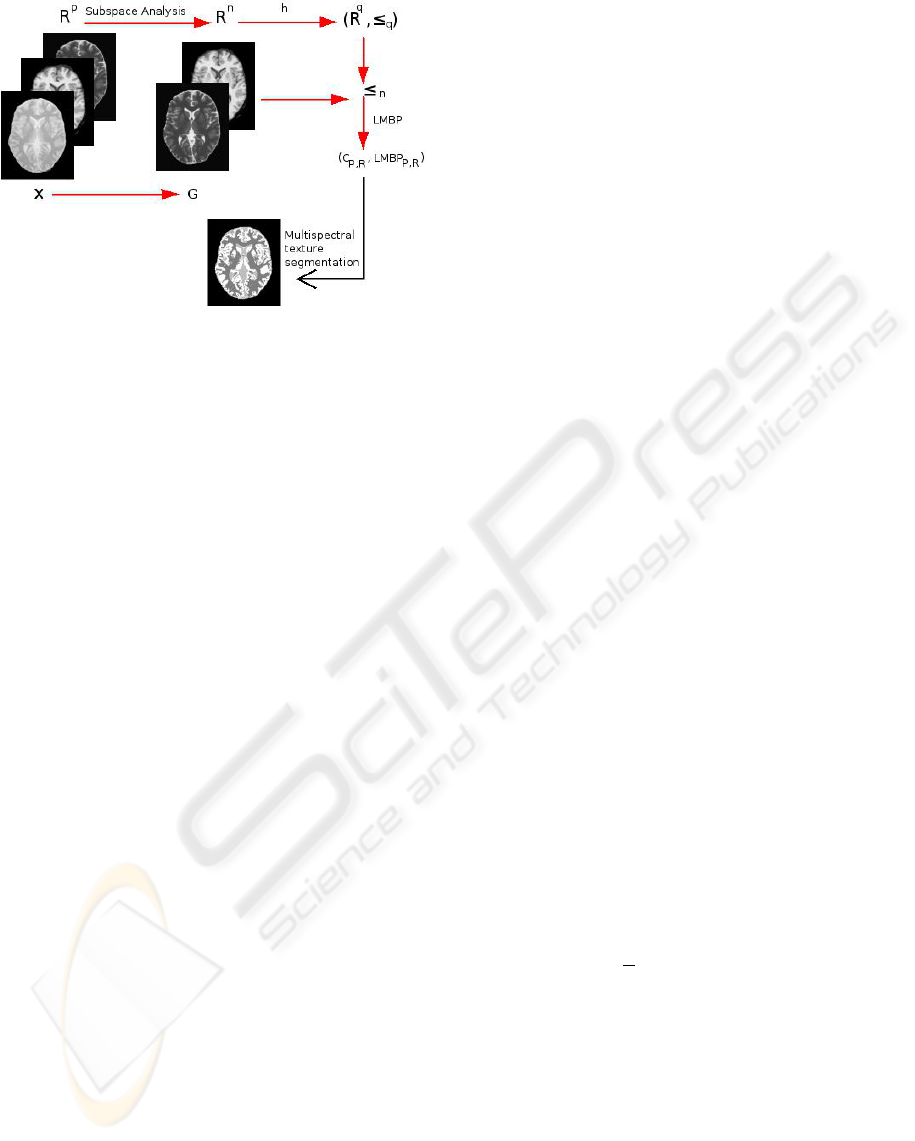
Figure 1: Overview of the algorithm.
features (Principal or Independent Component Anal-
ysis, Minimum Noise Fraction, or nonlinear tech-
niques such as Isomap or Local Linear Embedding).
In a preliminary study, we used a simple and linear
technique, the Principal Component Analysis (PCA),
that will directly impact the choice of the h function:
if X ∈ M
m,p
(R) denotes the matrix of the original
data (m being the number of pixels, p the number
of images and X
.,i
the i
th
column of X), the princi-
pal components are computed from the projections of
the original data onto the eigenvectors of Z
T
Z, where
Z = (X − 1µ)D is the matrix of centered and reduced
data, 1 = (1···1) ∈ R
m
,µ = (
¯
X
.,1
···
¯
X
.,p
) ∈ R
p
is the
vector of image mean values and D = diag(1/s
i
),
s
i
,1 ≤ i ≤ p is the standard deviation of variable i.
PCA allows n < p new variables to be computed, ex-
plaining most of the variance of the original data. The
resulting information is stored in G ∈ M
m,n
(R).
2.3.2 Definition of h
Recall that h : R
n
→ R
q
, an easily way to compute an
ordering relation in R
n
is to derive it from h using the
canonical ordering relation of R
q
,q ≥ 1:
x ≤
q
y ⇔ (∀1 ≤ i ≤ q, x(i) ≤ y(i))
This ordering relation is a partial order, and in the con-
text of image processing may lead to several draw-
backs: some vectors may not be ordered, and notions
of Sup and Inf are not defined (and are compulsory in
areas such as mathematical morphology). In our con-
text, we thus decide to define a total ordering in R
n
,
then using q = 1 and h injective. Several choices are
possible (e.g. the bit mixing approach (Chanussot and
Lambert, 1998)), and we benefit from the new basis
computed from PCA to use the lexicographic order in
R
n
defined as:
x ≤
n
y ⇔ (∃k ∈ {1 ·· ·n})/x(i) = y(i),1 ≤ i ≤ k − 1, x(k) < y(k)
If each image is coded on b bits, the corresponding h
function is
h(x) =
n
∑
i=1
x(n + 1 − i)2
b(i−1)
2.3.3 Computation of LMBP
Once h has been defined, an ordering relation on R
n
can be proposed as: x ≤
n
y ∈ R
n
⇔ h(x) ≤ h(y) ∈ R.
Unfortunately, enforcing a total ordering on R
n
makes
h discontinuous: the Netto theorem (Sagan, 1994)
indeed proved that any bijective application from a
manifold of dimension n to a manifold of dimension
q 6= n is discontinuous. Thus, h is not a linear func-
tion, and does not commute with linear functions.
It is thus not straightforward to assess h(x) − h(y),
given x − y in R
n
, and any linear combination of vec-
tors transformed by h should be avoided. If this is
not really a problem for the definition of the LBP
operator (σ(g
p
− g
c
) should easily be replaced with
σ(h(g
p
) − h(g
c
)) because the minus sign only means
to compare h(g
p
) with h(g
c
) using the ordering de-
fined by h), the extension of C
P,R
to the case of multi-
spectral images is more problematic, because of both
¯g and the deviation to this mean value. In order to be
compliant with the discontinuity of h, we thus avoid
any linear combination, and replace ¯g by the median
value m
p
of the g
p
’s. The LBMP operator is then
LMBP
riu2
P,R
=
P−1
∑
i=0
σ(h(g
p
) − h(g
c
)) if U(LBP
P,R
) ≤ 2
P + 1 otherwise
where
U(LBP
P,R
) = |σ(h(g
P−1
) − h(g
c
)) − σ(h(g
0
) − h(g
c
))|
+
P−1
∑
i=1
|σ(h(g
i
) − h(g
c
)) − σ(h(g
i−1
) − h(g
c
))|
and the contrast operator
C
P,R
=
1
P
P−1
∑
i=0
(h(g
i
) − h(m
p
))
2
C
P,R
is still a combination of h(g
i
) − h(m
p
), but we
expect the nonlinearity introduced by m
p
to reduce
the discontinuity effects.
If h is computed from the lexicographic order, the sign
functions σ((h(x)−h(y))) in the previous expressions
reduce to
σ(
n
∑
j=1
(x(n + 1 − j) − y(n + 1 − j))2
b( j−1)
)
The first components of x and y play here an impor-
tant role, as their difference is weighted by 2
b(n−1)
,
MULTISPECTRAL TEXTURE ANALYSIS USING LOCAL BINARY PATTERN ON TOTALLY ORDERED
VECTORIAL SPACES
39

but the other components, with weights 2
b. j
, j < n − 1
may invert the sign of the argument of the σ function.
Subspace analysis is thus a crucial step in the whole
process for the selection of relevant components.
2.3.4 From LMBP to Segmentation
Once LMBP and contrast have been computed for
each pixel location, we were interested in image seg-
mentation using these features. Several approaches
can be performed, e.g.:
• incorporate these operators in a vector describ-
ing the pixel properties, with other relevant val-
ues (e.g. values of the first principal components).
The set of these vectors then serves as an input of
an unsupervised clustering algorithm
• compute a local 2D joint distribution (LMBP,C
P,R
)
for each pixel, and use an adapted metric to cluster
pixels.
In this preliminary study, we chose to use the first al-
ternative. More precisely, a classical K-means algo-
rithm was used as a clustering method, using the Eu-
clidean metric to cluster feature vectors that the next
section will detail.
3 RESULTS
We apply the LMBP operator to the problem of mul-
tispectral MR image segmentation problem. As test
data we used simulated MRI-datasets generated with
the Internet connected MRI Simulator at the Mc-
Connell Brain Imaging Centre in Montreal
(www.bic.mni.mcgill.ca/brainweb/). The datasets we
used were based on an anatomical model of a normal
brain that results from registering and preprocessing
27 scans from the same individual with subsequent
semi-automated segmentation. In this dataset the dif-
ferent tissue types were well-defined, both “fuzzy”
and “crisp” tissue membership were allocated to each
voxel. From this tissue labeled brain volume the MR
simulation algorithm, using discrete-event simulation
of the pulse sequences based on the Bloch equations,
predicted signal intensities and image contrast in a
way that is equivalent to data acquired with a real
MR-scanner. Both sequence parameters and the effect
of partial volume averaging, noise, and intensity non-
uniformity were incorporated in the simulation results
(Cocosco et al., 1997; Kwan et al., 1999).
Ten multispectral (T1-weighted,T2-weighted, Proton
density) MR datasets of a central slice (including the
main brain tissues, basal ganglia and fine to coarse
details), with variations of the parameters ”noise” and
”intensity non-uniformity (RF)” were chosen (table
1), the slice thickness being equal to 1mm. This se-
lection covers the whole range of the parameter values
available in BrainWeb so that the comparability with
real data can be considered as sufficient to test the ro-
bustness of the method at varying image qualities.
Table 1: MR Datasets.
dataset no dataset name noise RF
1 n1rf20 1% 20%
2 n1rf40 1% 40%
3 n3rf20 3% 20%
4 n3rf40 3% 40%
5 n5rf20 5% 20%
6 n5rf40 5% 40%
7 n7rf20 7% 20%
8 n7rf40 7% 40%
9 n9rf20 9% 20%
10 n9rf40 9% 40%
For obtaining the true volumes of brain tissues and
background the corresponding pixels were counted in
the ground truth image provided by BrainWeb.
We performed three types of analysis for each dataset,
first transformed by a subspace analysis method
(namely the PCA). More precisely, we characterized
pixels with several types of feature vectors:
• either the vector of the first principal components,
or a vector composed of the first principal compo-
nents, the LMBP and the contrast operators (In-
terest of LMBP and Multispectral Contrast in the
segmentation process, section 3.1).
• either a vector composed of the first principal
components, the LMBP and the contrast oper-
ators, or a vector composed of the first princi-
pal components, the multispectral and the contrast
operators as computed in (Song et al., 2006) (sec-
tion 3.2)
In the following, we present results from dataset
n9rf20 (figure 2) and use as LBP parameters R =
1.5,P = 12. All features were normalized by their
max value to cluster homogeneous values.
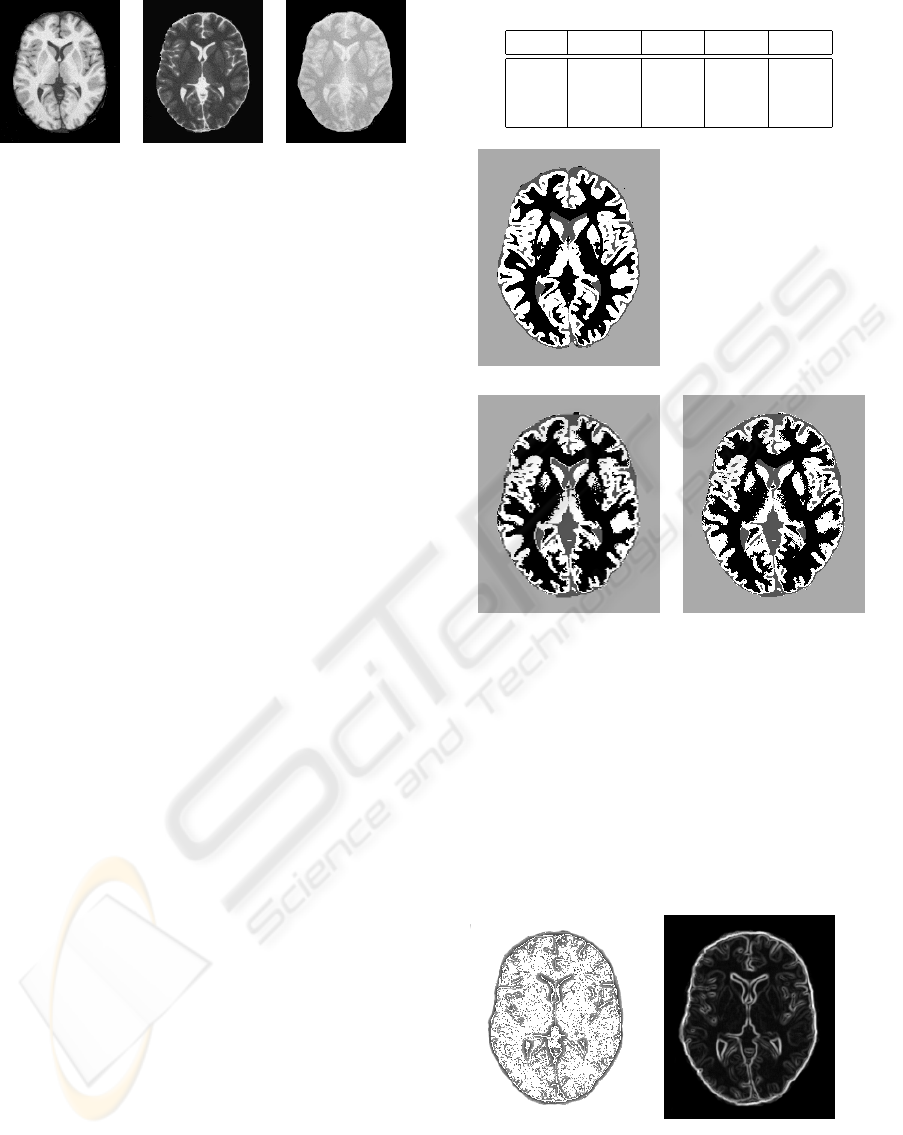
3.1 Multispectral Texture Information
Figure 3 presents the results of the segmentation of
the brain slice in 4 classes: background (BG, light
gray), Cerebrospinal fluid (CSF, dark gray), white
matter (WM, black) and gray matter (GM, white). S1
is the segmentation obtained with only the two first
principal components, S2 with these two components
plus the LMBP and the contrast operator values.
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
40
T1-weighted T2-weighted Proton Density
Figure 2: Slice of interest of dataset n9rf20.
In order to assess the two segmentations, we pro-
cess the confusion matrix C on the segmented images:
given a segmented image S, C
S
(i, j) is the percentage
of pixels assigned to class i ∈ {BG,CSF,W M, GM} in
the ground truth and to class j in S. The closer C to
a diagonal matrix, the better the segmentation is. Al-
though segmented images seems to be the same, the
confusion matrix reveal some differences:
C
S1
=
48.4 10
−1
1.8.10
−3
0
0.44 4.64 1.58 0.83
0.06 0.12 19.28 4.62
0.13 0.89 0.71 18.2
C
S2
=
48.3 0.10 2.10
−2
0
0.46 5.73 0.56 0.88
0.05 0.05 19.28 4.63
0.13 0.87 0.45 18.45
The multispectral texture coefficients, and espe-
cially the LMBP operator, bring some information on
structural properties on tissue edges: interfaces be-
tween tissues are more or less expressed depending on
the acquisition (e.g. the CSF/(WM+GM) interface is
very clear in T2-weighted images, whereas the proton
density images better shows the WM/(GM+CSF) in-
terface), and the multispectral structure of edges, seen
as local oriented textures, is managed during the clus-
tering process (figure 4). It is particularly visible for
the WM/CSF interface between the corpus callosum
and the lateral ventricles (see image and C(2, 3) coef-
ficient) and for the WM/GM interface near the puta-
mens (see image and C(4,3) coefficient).
Table 2 shows the number of pixels assigned to
each class. Relative errors for S1 (respectively S2)
are 0.01 (0.01) for background, 0.31 (0.31) for CSF,
0.11 (0.06) for WM and 0.15 (0.11) for GM. For WM
and GM, relative errors lower due to the integration
of LBMP operators.
3.2 LMBP vs. Another Multispectral
Texture Definition
We also compared the LMBP operator with a mul-
tispectral LBP already proposed in the litterature
(Song et al., 2006), that compute LBP as:
Table 2: Number of pixels in each class.
BG CSF WM GM
truth 19826 2336 8741 9577
S1 19620 3039 9755 8066
S2 19601 3088 9328 8463
Ground truth
S1 S2
Figure 3: Segmentation results.
LBP
riu2
P,R
=
P−1
∑
i=0
σ(kg
p
k − kg
c
k) if U(LBP
P,R
) ≤ 2
P + 1 otherwise
where norms also stand in the definition of U. As
in (Song et al., 2006), we used the Euclidean norm,
and the LBP parameters were chosen equal to those
of LMBP. Note that this method is a particular case of
LMBP, with h : x 7→ kxk.
LBMP Contrast
Figure 4: LBMP and Contrast images.
Figure 5 shows LMBP and Contrast images for
both methods, and figure 6 compares the two seg-
MULTISPECTRAL TEXTURE ANALYSIS USING LOCAL BINARY PATTERN ON TOTALLY ORDERED
VECTORIAL SPACES
41
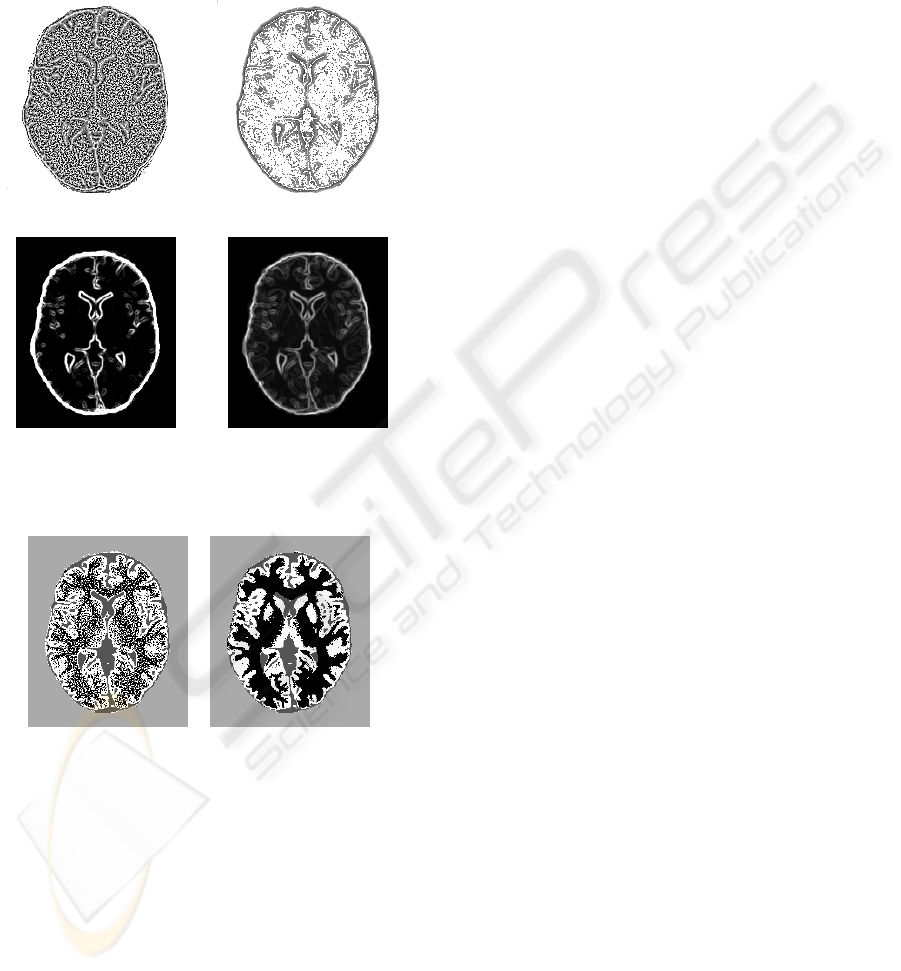
mentations. Results were always much better using
LMBP for all the MR volumes described in Table 1,
and the difference increased as the noise increased in
the image. The function h : x 7→ kxk used to produce
the total ordering in R
n
indeed tended to increase the
lbp value in a noisy neighborhood of a pixel g
c
, pro-
ducing the noisy LBP image of figure 5.
(Song et al., 2006) LBP LMBP
(Song et al., 2006) contrast LMBP Contrast
Figure 5: Comparison of multispectral LBP and Contrast
images.
(Song et al., 2006) LMBP segmentation
segmentation
Figure 6: Comparison of segmentation results.
4 CONCLUSIONS
We proposed in this article a multispectral version of
the classical Local Binary Pattern operator, based on
a total ordering on the vectorial space of the data. We
demonstrate its efficiency on multispectral MR im-
ages of the brain, assessing the results with respect to
a ground truth, and comparing segmentation results
with those provided by another multispectral LBP ap-
proach.
Numerous perspectives are now expected from this
preliminary work. First of all, h needs to be better
defined to allow the local topology to be preserved:
two neighbors in R
n
need to stay closed when trans-
formed by h. For x and y neighbors in R
n
, the solu-
tion may be to define h using space filling curves di-
rectly on the multispectral image, in order to impose
small variations of h(x)−h(y) in areas of interest, and
higher variations for example in the background.
The segmentation scheme also needs to be refine. For
this study, standard techniques (PCA, Kmeans) were
applied, and some work has now to be done to tune
subspace analysis and segmentation methods to this
specific problem. Finally, this multispectral approach
finds natural applications not only in medical imag-
ing, but also in remote sensing imagery. We now in-
tend to tune and apply the LMBP to this domain.
REFERENCES
Ahonen, T., Hadid, A., and Pietik
¨
ainen, M. (2006). Face
description with local binary patterns: Application to
face recognition. IEEE Trans. Pattern Anal. Mach.
Intell., 28(12):2037–2041.
Barnett, V. (1976). The ordering of multivariate data. Jour-
nal of the Royal Statistical Society, Series A, 139:318–
355.
Chanussot, J. and Lambert, P. (1998). Total ordering based
on space filling curves for multivalued morphology.
In ISMM ’98: Proceedings of the fourth international
symposium on Mathematical morphology and its ap-
plications to image and signal processing, pages 51–
58, Norwell, MA, USA. Kluwer Academic Publish-
ers.
Cocosco, C. A., Kollokian, V., Kwan, R. K. S., Pike, G. B.,
and Evans, A. C. (1997). Brainweb: Online interface
to a 3d mri simulated brain database. NeuroImage, 5.
Garc
´
ıa, M. A. and Puig, D. (2007). Supervised texture
classification by integration of multiple texture meth-
ods and evaluation windows. Image Vision Comput.,
25(7):1091–1106.
Goutsias, J. K., Heijmans, H. J. A. M., and Sivakumar,
K. (1995). Morphological operators for image se-
quences. Computer Vision and Image Understanding,
62(3):326–346.
Heikkil
¨
a, M., Pietik
¨
ainen, M., and Schmid, C. (2009). De-
scription of interest regions with local binary patterns.
Pattern Recogn., 42(3):425–436.
Kwan, R., Evans, A., and Pike, G. (1999). Mri simulation-
based evaluation of image-processing and classifica-
tion methods. 18(11):1085–1097.
Lucieer, A., Tsolmongerel, O., and Stein, A. (2005). Multi-
variate texture-based segmentation of remotely sensed
images. International Journal of Remote sensing,
26:2917–2936.
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
42

M
¨
aenp
¨
a
¨
a, T. and Pietik
¨
ainen, M. (2003). Multi-scale binary
patterns for texture analysis. pages 885–892.
Ojala, T., Pietikinen, M., and Menp, T. (2002). Multires-
olution gray-scale and rotation invariant texture clas-
sification with local binary patterns. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
24(7):971–987.
Paclik, P., Duin, R. P., Kempen, G. M. P. V., and Kohlus,
R. (2002). Supervised segmentation of textures in
backscatter images. In In: Proceedings of IEEE In-
ternational Conference on Pattern Recognition, pages
490–493. John Wiley and Sons.
Sagan, H. (1994). Space Filling Curves. Springer Verlag.
Song, C., Li, P., and Yang, F. (2006). Multivariate texture
measured by local binary pattern for multispectral im-
age classification. In In: Proceedings of IEEE Inter-
national Conference on Geoscience and Remote Sens-
ing Symposium, pages 2145–2148.
Tuceryan, M. and Jain, A. K. (1998). Texture analysis, vol-
ume 15 of Handbook of Pattern Recognition and Com-
puter Vision, Second Edition, pages 207–248. World
Scientific, c.h. chen, and l.f. pau edition.
Wang, A. P. and Wang, S. G. (2006). Content-based high-
resolution remote sensing image retrieval with local
binary patterns. In Society of Photo-Optical Instru-
mentation Engineers (SPIE) Conference Series, vol-
ume 6419 of Society of Photo-Optical Instrumentation
Engineers (SPIE) Conference Series.
MULTISPECTRAL TEXTURE ANALYSIS USING LOCAL BINARY PATTERN ON TOTALLY ORDERED
VECTORIAL SPACES
43
