
The Impact of Pre-processing on the Classification of
MEDLINE Documents
Carlos Adriano Gonc¸alves
1
, C
´
elia Talma Gonc¸alves
2,3
, Rui Camacho
1
and Eug
´
enio Oliveira
2
1
LIAAD & FEUP - Faculdade de Engenharia da Universidade do Porto, Porto, Portugal
2
LIACC & FEUP - Faculdade de Engenharia da Universidade do Porto, Porto, Portugal
3
CEISE & ISCAP - Instituto Superior de Contabilidade e Administrac¸
˜
ao, Porto, Portugal
Abstract. The amount of information available in the MEDLINE database
makes it very hard for a researcher to retrieve a reasonable amount of relevant
documents using a simple query language interface. Automatic Classification of
documents may be a valuable technology to help reducing the amount of docu-
ments retrieved for each query. To accomplish this process it is of capital impor-
tance to use appropriate pre-processing techniques on the data. The main goal of
this study is to analyse the impact of pre-processing techniques in text Classifi-
cation of MEDLINE documents. We have assessed the effect of combining dif-
ferent pre-processing techniques together with several classification algorithms
available in the WEKA tool. Our experiments show that the application of prun-
ing, stemming and WordNet reduces significantly the number of attributes and
improves the accuracy of the results.
1 Introduction
Molecular biology and biomedicine scientific publications are available (at least the
abstracts) in Medical Literature Analysis and Retrieval System On-line (MEDLINE).
MEDLINE is the U.S. National Library of Medicine’s (NLM) premier bibliographic
database that contains over 16 million references to journal articles in life sciences with
a concentration on biomedicine. A distinctive feature of MEDLINE is that the records
are indexed with NLM’s Medical Subject Headings (MeSH terms). MEDLINE is the
major component of PubMed [1], a database of citations of the National Library of
Medicine in the United States. PubMed comprises more than 19 million citations for
biomedical articles from MEDLINE and life science journals. The result of a MED-
LINE/PubMed search is a list of citations
4
to journal articles. The results of such search
is, quite often, a huge amount of documents, making it very hard for researchers to ef-
ficiently reach the most relevant documents for their queries. As this is a very relevant
and actual topic of investigation we investigate the use of Machine Learning-based text
classification techniques to help in the identification of a reasonable amount of rele-
vant documents in MEDLINE. In this study we will focus on assessing the effect of
different pre-processing techniques in the quality of classifiers available in the Weka
tool.
4
including authors, title, journal name, the abstract of the paper, keywords and MeSH terms
Adriano GonÃ
˘
galves C., Talma GonÃ
˘
galves C., Camacho R. and Oliveira E.
The Impact of Pre-processing on the Classification of MEDLINE Documents.
DOI: 10.5220/0003028700530061
In Proceedings of the 10th International Workshop on Pattern Recognition in Information Systems (ICEIS 2010), page
ISBN: 978-989-8425-14-0
Copyright
c
2010 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

The rest of this paper is structured as follows. In Section 2 we overview the pre-
processing techniques used. In Section 3 we present the text classification problem and
existing techniques. We report on related work in Section 4. Section 5 describes the ex-
periments and Section 6 the results obtained. Section 7 concludes the paper presenting
the conclusions and future work.
2 Pre-processing Techniques
Before applying any data analysis technique it is necessary to pre-process the collection
of documents. In our study we have used the following pre-processing techniques:
– Tokenization, the process of breaking a text into tokens. A token is a non-empty
sequence of characters, excluding spaces and punctuation.
– Lowercase conversion converts all terms into lower cases.
– Special character removal removes all the special characters (+, -, !, ?, ., ,, ;, :, {,
}, =, &, #, %, $, [, ], /, <, >, \, “, ”, |) and digits.
– Stop Word Removal removes words that are meaningless such as articles. con-
junction and prepositions (e.g., a, the, at, etc.). These words are meaningless for the
evaluation of the document content.
– Stemming is a widely used technique in text analysis. Stemming is the process of
removing inflectional affixes of words reducing the words to their stem.
– Pruning discards terms either appearing rarely or “too frequently”. Terms that
rarely appear in a document or terms that appear too frequently do not contribute
to identify the topic of the document. The most common techniques are term fre-
quency and document frequency.
– Treating synonyms: the possibility to take care of synonyms may be seen as an-
other pre-processing technique. If two words or terms mean the same thing, e.g, if
they are synonyms we could replace them by one of them without taking the se-
mantic meaning of the term.
– Document representation: after the above listed “filters”, each document is en-
coded and stored as a standard vector of term weights.
In the Vector Space Model there are several variations to attribute the weights to the
terms. Two of the most common weights are: TF (Term Frequency) and TFIDF (Term
Frequency Inverse Term Frequency). We have used TFIDF in our study. [2] provides
the following definitions:
T F (term, document) = thefrequencyof termindocument (1)
54

and
IDF = log
numberofdocumentsincollection
numberofdocumentswithterm
+ 1. (2)
These pre-processing techniques are of utmost importance since they reduce sig-
nificantly the number attributes that characterise each document attenuating the curse
of dimensionality. Stop word removal, stemming and pruning improves classification
quality once they remove the meaningless data that leads to a reduction in the num-
ber of dimensions in the term space. Document representation concerns the estimation
of the importance of a specific term in the document. As the number of features (at-
tributes) are very huge in a collection of documents, the pre-processing techniques help
in reducing these huge number of features.
3 Text Classification
Text Classification attempts to automatically determine whether a document or part of a
document has particular characteristics of interest, usually based on whether the docu-
ment discusses a given topic or contains a certain type of information [3]. Text Classifi-
cation involves two main research areas: Information Retrieval and Machine Learning.
The first of step of Text Classification is to transform documents into a suitable rep-
resentation for the Classifier. For this, and before applying the Classifier documents
must be pre-processed using Information Retrieval techniques mentioned in the previ-
ous section. Some other pre-processing techniques that can be applied to the collection
of documents, in order to reduce the huge amount of terms in the collection is called
Feature Selection.
A Feature Selection or feature extraction phase is needed to reduce the dimensionality
of the document.
There are three Classification techniques: supervised learning, unsupervised learning
and semi-supervised learning.
Supervised Learning is based on a training set of examples, where the key idea is to
learn from a set of labelled examples (the training set).
In unsupervised learning there is no a priori output. The goal of unsupervised learning
is to learn a model that explains well the data. Usually, the result of unsupervised learn-
ing is a new explanation or representation of the observation data, which will then lead
to improved future decisions.
Semi-supervised learning makes use of both labelled and unlabelled data (typically a
small amount of labelled data and a large amount of unlabelled data. Semi-supervised
learning [4] [5] is a machine learning paradigm in which the model is constructed with
a small number of labelled instances and a large number of unlabelled instances. One
key idea in semi-supervised learning is to label unlabelled data using certain techniques
and thus increase the amount of labelled training data.
4 Related Work
The authors in [6] explore the effects of different text representation approaches on the
classification performance of MEDLINE documents. In this study the authors use only
55

the title and combine a word representation, the bag-of-words approach with a phrase
representation, the bag-of-phrase. Due to previous studies [7] they decided to compare
this approach with the bag-of-words representation, bag-of-phrase representation and
an hybrid one. They also used the Support Vector Machine Algorithm. The experiments
were made with OHSUMED
5
data set. The authors achieved better performance results
with the hybrid approach.
In [8] the authors examined ways to represent text from two aspects related with
text representation in a vector space model: (1) what should a term be and (2) how to
weight a term. The authors evaluated their approach using a Support Vector Machine
algorithm. They found that representing text using a different approaches from the bag-
of-words representation does not show performance improvements. They also found
that the term weight slightly improved the performance.
In this paper we evaluate the impact of pre-processing techniques ( stop word re-
moval, stemming, WordNet and pruning ) in Classification, that as far as our knowledge
have not yet been studied (concerning a MEDLINE data set).
5 Empirical Study
Our base of work will be a MEDLINE sample. The focuses of this research is to study
the impact of pre-processing techniques in a MEDLINE sample Data Set.
5.1 Data Set Characterisation
The data set that is subject of our study, is a MEDLINE sample that was downloaded
from the NLM site at (ftp://ftp.nlm.nih.gov/nlmdata/sample/MEDLINE/). This sample
has 53.2 MB, and contains 30000 citations. The sample is in the XML format. Each
citation contains several information namely: the pmid (the PubMed id), the journal
title, the PubMed date, the article title, the abstract of the paper if available, the list
of authors, the list of keywords and the list of Mesh terms. A MeSH (Medical Subject
Headings) is the (U.S.) National Library of Medicine’s controlled vocabulary thesaurus.
It consists of sets of terms naming descriptors in a hierarchical structure that permits
searching at various levels of specificity. A MeSH term is a medical subject heading,
or descriptor, as defined in the MeSH thesaurus. We have made a pre-selection of 1098
papers using only the one’s containing at least one of the following mesh headings:
“Erythrocytes”, “Escherichia coli”, “Protein Binding” and “Blood Pressure” containing
approximately 250 documents per category.
5.2 Pre-processing Techniques Applied
The MEDLINE sample we used in our study was in the XML format. So the first pre-
processing step was to read the XML file and to filter the information we need namely
the pmid (PubMed id), journal title, PubMed date, article title, abstract of the paper if
5
which is a database composed of 348,566 MEDLINE documents from 270 journals that are
classified under 14,321 mesh categories
56

available, list of authors, list of keywords and the list of Mesh terms. We have devel-
oped our application using the JAVA Programming Language which contains classes to
work with XML files and SQL, that are needed for our approach. We have also used
a MySQL Database to store all the information useful for further pre-processing and
Classification. We have applied the following pre-processing techniques to our original
Data Set.
– Tokenization;
– Stop Words removal: we have used a set of 659 stop words;
– We have used WordNet [9] to search for synonyms of terms and replace each term
for the respective synonym (we have chosen the smallest one);
– Stemming: we used the Porter’s Stemmer Algorithm [10]
– We have implemented the standard term-frequency inverse document frequency
(TFIDF) function to assign weights to each term in the document.
Besides this most common pre-processing techniques we have applied some other
pre-processing features with the objective of reducing the number of attributes, namely:
– Pruning:
• We have removed bi-grams words that are meaningless (such as “iz”, “tk”,
etc.);
• We have also removed the words that appear less than 10 times in the all collec-
tion, because if their frequency is so low probably they are not discriminative
of the document content;
• We have also removed the terms that do not appear in the WordNet neither on
a medical dictionary of terms (The Hosford Medical Terms Dictionary [11]).
The main reason is to eliminate terms that are not valid (such as “tksx”) that
are meaningless and do not constitute a valid term.
As already mentioned we have used the TFIDF method because in this weighting
scheme terms that appear too rarely or too frequently are ranked lower than terms that
balance between the two extremes. And also because higher weight terms signify that
the term contributes better to classification results.
Figure 1 summarises our approach.
5.3 Data Warehouse Techniques
Data Warehouse “is a subject-oriented, integrated, non- volatile and time-variant col-
lection of data in support of management’s decisions ” as defined by [12]. Data Ware-
house techniques are used to represent the collection of documents as a set of vectors
that can be written as a matrix. A dimensional model comprises facts, dimensions and
measures [13]. The fact table represents the measure that is being tracked [14]. In our
study was used a Snowflake schema with a central fact table of terms. The ETL process
(Extracting, Transforming and Loading and Indexing) was made using a SQL database.
The Extract was made using the XML source files and populating the SQL database.
The Fact Term table contains measures like the number of occurrence of the term at the
title and also at the abstract (representing the T F = Term Frequency). Other measures
57
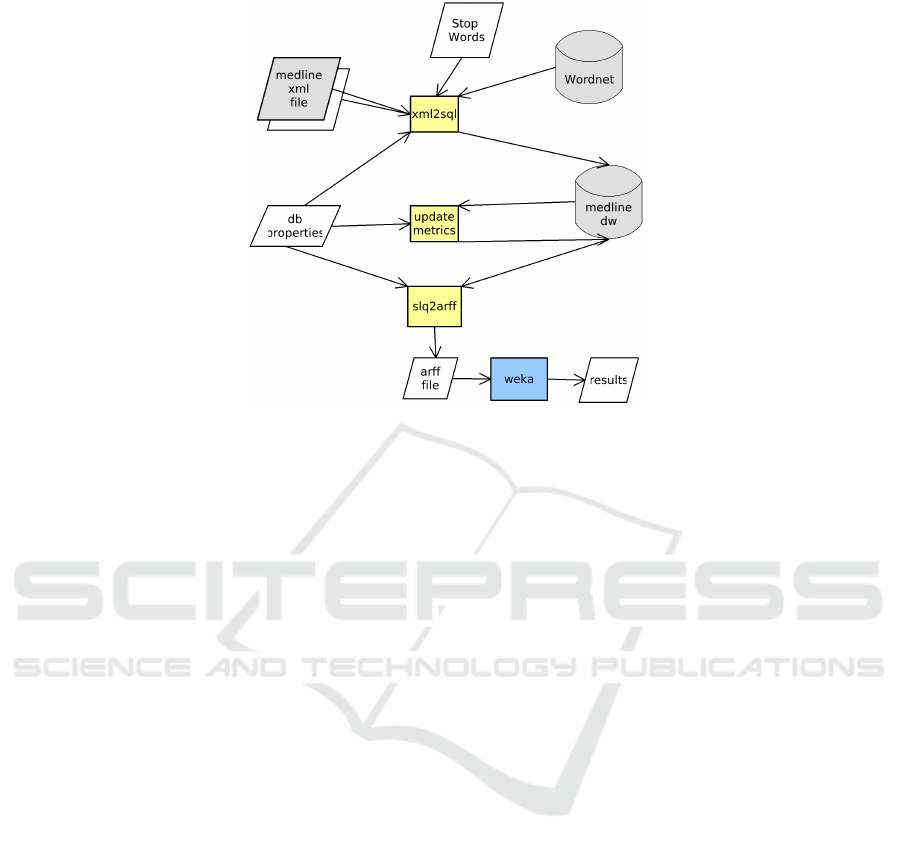
Fig. 1. Summary of steps.
introduced in the fact table was the T F ∗IDF (Term Frequency Inverse Document Fre-
quency) to calculate the “Weight” of a term in a document. To calculate these measures,
another measure must be used - the Document Frequency - representing the frequency
of the term in all the documents collection. This measure can be calculate using the
dimension term table. Our model (summarised at Figure 2) includes other dimension
tables like a document dimension to represent the citation document; a Time dimension
was used to represent the time dimension associated with the citation document; author
dimension that have to be represented using a group author due to the M ←→ M rela-
tion between the term fact table and the authors dimension. Using this techniques, the
calculation process that is represented by the “update metrics” in Figure 1 perform the
final calculations and was made using only SQL instructions. With this representation,
the creation of the data set is simplified, since it can be done using SQL instructions to
join the records, according to our needs to produce the output data set. Also, the pruning
can be made with SQL instructions that limit the number of terms that should be used.
The last step is represented by the “sql2arff” process in Figure 1 and was applied the
following pruning technique through our Data Warehouse:
– < 10∗ To remove words that appear less than 10 times in the document collection.
– < 100∗ To remove words that appear less than 100 times in the document collec-
tion.
All the SQL instruction used by our Java programs are on the db.properties file in
Figure 1. If we decide to change the Mesh, the pruning to apply or any other character-
istics we simple need to change this file.
With this technique was possible to reduce the number of attributes from more than
1300 to less then 90. This can have a significantly impact in processing the MEDLINE
database.
58
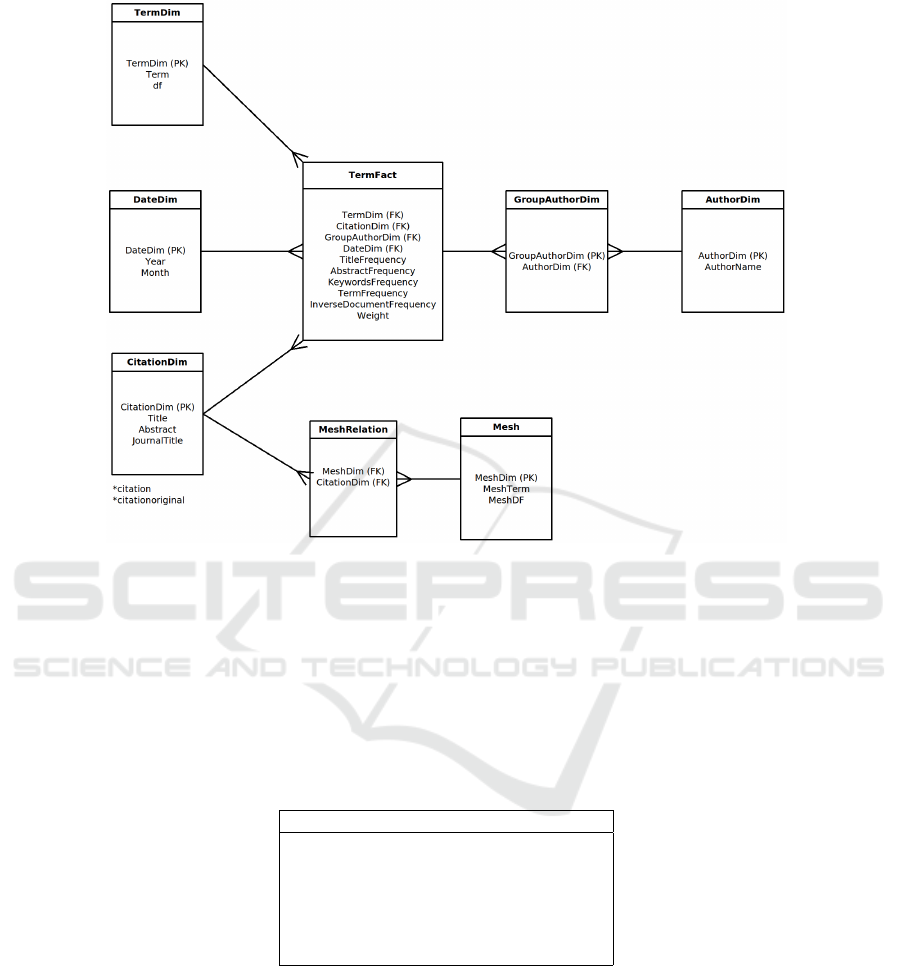
Fig. 2. Snowflake Schema.
6 Experimental Results
Table 1 lists the algorithms available in WEKA and used in our experiments. We have
generated several data sets combining the different pre-processing techniques (pruning,
stemming and WordNet). The quality of the classifiers were assessed by their Accuracy.
Table 1. Machine Learning algorithms used in the study. RF stands for Random Forest.
Algorithm Type
SMO Support Vector Machine
RF Ensemble algorithm
IBk K-nearest neighbours
BayesNet Bayes Network
j48 Decision tree
dtnb Decision table/naive bayes hybrid
The results obtained are in Table 2. In the Pruning column we present the thresholds
for term occurrence (terms that appear less than 10 and less than 100 times (df) in the
document collection and that were removed).
The first experiences shows that the application of pruning, stemming and WordNet
reduces significantly the number of attributes without affecting the accuracy of results.
59
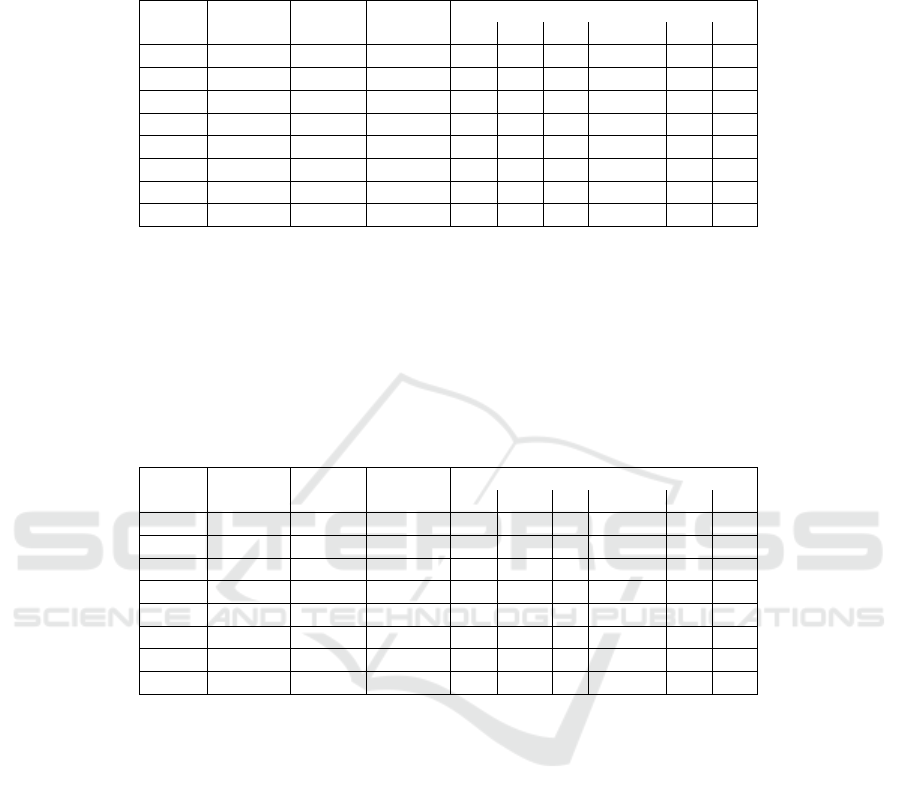
Table 2. Accuracy Classification Results. The majority class is 28%.
Pruning Stemming WordNet Attributes Acc
SMO RF IBk BayesNet j48 dtnb
< 10∗ no no 1304 76.95 84.09 46.66 86.85 81.33 76.28
< 100∗ no no 62 76.62 81.70 63.02 81.60 80.74 77.01
< 10∗ yes no 1103 80.57 84.85 49.71 86.85 84.57 80.57
< 100∗ yes no 90 77.31 85.51 61.10 85.03 84.17 79.88
< 10∗ no yes 916 76.38 84.95 48.19 87.14 83.90 79.42
< 100∗ no yes 85 80.24 85.20 61.45 85.97 83.77 79.77
< 10∗ yes yes 904 78.00 85.14 51.42 87.42 83.71 80.66
< 100∗ yes yes 91 77.71 85.80 57.33 86.38 82.66 80.47
We get the best results using the Random Forest (85.80%) using a pruning of df = 100,
using the stemming and the WordNet. The accuracy result are even best with only 91
terms than using 904 terms. The Bayes Network algorithm with a pruning of df = 10 ,
with Stemming and WordNet show the best result, but if we look at the result of using
pruning with df = 100 the accuracy drop from 87.42% to 86.38%.
The time taken to build the models are presented on in Table 3.
Table 3. Time to construct the models in seconds.
Pruning Stemming WordNet Attributes Time
SMO RF IBk BayesNet j48 dtnb
< 10∗ no no 1304 1.32 101.98 0.01 2.5 8.16 34.05
< 100∗ no no 62 0.8 2.39 0.01 0.38 0.56 3.19
< 10∗ yes no 1103 1.27 69.69 0.01 2.03 10.55 54.64
< 100∗ yes no 90 0.75 2.39 0.02 0.43 1.17 5.96
< 10∗ no yes 916 1.45 61.23 0.01 1.74 7.81 58.78
< 100∗ no yes 85 1.25 3.14 0.01 0.8 1.4 3.68
< 10∗ yes yes 904 1.77 59.97 0.01 1.6 8.16 34.05
< 100∗ yes yes 91 1.3 3.96 0.04 0.74 1.24 5.56
The time taken to build the models were obtained on a intel Core 2 Duo Processor
@ 2.40Ghz with 4096 GB. We can see that pruning has a positive impact in the time
taken to build the models specially on algorithms that take longer to execute.
7 Conclusions and Future Work
This paper focuses on the study of the impact of pre-processing techniques in classi-
fying MEDLINE documents. We have presented results of our empirical study of the
impact of the pre-processing techniques in text classification. The amount of informa-
tion available on the MEDLINE database can (and probably will) be an issue. Through
the use of Information Retrieval and Data Warehouse techniques applied it was possi-
ble to reduce significantly the time needed for the pre-processing without affecting the
accuracy. Although we are using a MEDLINE sample we have a data set with 30000
MEDLINE citations and we selected only 1098 citations with abstract representing
60

the 4 classes using only the one’s containing at least one of the following mesh head-
ings: “Erythrocytes”, “Escherichia coli”, “Protein Binding” and “Blood Pressure”. In
this sample we started with more than 1300 attributes that can be represented using a
fact table and some dimensions tables in our Snowflake schema data warehouse. The
pruning pre-processing was made using only SQL instructions on the Data Warehouse.
Unlike [6] we also have used the abstract from the papers. Like [8] we have used a
bag-of-words representation and we implemented the standard term-frequency inverse
document frequency (TFIDF) function to assign weights to each term. We have gener-
ated several data sets combining the different pre-processing techniques. We have made
a comparison table of the accuracy obtained using different pre-processing techniques
and different classification algorithms. We have also presented the computation time re-
quired for the execution of the algorithms. The best accuracy result achieved (87.42%)
is thus promising.
As a future work and to achieve a better classification we may: 1) incorporate more
information 2) optimise the MeSH terms selection for each document 3) test with other
MEDLINE sources, like the 2010 MEDLINE version and 4) using the full MED-
LINE database.
References
1. Wheeler, D. L., Barrett, T., Benson, D. A., Bryant, S. H., Canese, K., Chetvernin, V., Church,
D. M., Dicuccio, M., Edgar, R., Federhen, S., Geer, L. Y., Helmberg, W., Kapustin, Y., Ken-
ton, D. L., Khovayko, O., Lipman, D. J., Madden, T. L., Maglott, D. R., Ostell, J., Pruitt,
K. D., Schuler, G. D., Schriml, L. M., Sequeira, E., Sherry, S. T., Sirotkin, K., Souvorov,
A., Starchenko, G., Suzek, T. O., Tatusov, R., Tatusova, T. A., Wagner, L., Yaschenko, E.:
Database resources of the national center for biotechnology information. Nucleic Acids Res
34 (2006)
2. Zhou, W., Smalheiser, N.R., Yu, C.: A tutorial on information retrieval: basic terms and
concepts. Journal of Biomedical Discovery and Collaboration 1 (2006)
3. Cohen, A.M., Hersh, W.R.: A survey of current work in biomedical text mining. Briefings
in Bioinformatics 6 (2005) pp. 57–71
4. Chapelle, O., et al.: Semi-supervised learning. Cambridge MIT Press (2006)
5. Zhu, X.: Semi-supervised learning literature survey (2006)
6. Meliha Yetisgen-Yildiz, W.P.: The effect of feature representation on medline document
classification. AMIA Annu Symp Proc. (2005) 849–853
7. Sebastiani, F.: Machine learning in automated text categorization. Computing Surveys. 31(1)
(2002) 1–47
8. LAN, M., TAN, C.L., SU, J., LOW, H.B.: Text representations for text categorization : a
case study in biomedical domain. In: IJCNN
ˆ
a07 : International Joint Conference on Neural
Networks. (2007)
9. Miller, G.A.: Wordnet: A lexical database for english. Communications of the ACM 38
(1995) pp. 39–41
10. Porter, M.F.: An algorithm for suffix stripping (1997)
11. Hosford medical terms dictionary v3.0 (2010)
12. Inmon, W.: Building the Data Warehouse. . (2002)
13. Schonbach, C., Kowalski-Saunders, P., Brusic, V.: Data warehousing in molecular biology.
Briefings in Bioinformatics 1 (2000) 190–198
14. Chituc, C.M.: Data warehousing - lecture notes (2009)
61
