
POPULATING BIOMEDICAL ONTOLOGIES FROM NATURAL
LANGUAGE TEXTS
Juana María Ruiz-Martínez, Rafael Valencia-García, Rodrigo Martínez-Béjar
Computer Science Faculty, University of Murcia, Campus de Espinardo, 30100 Murcia, Spain
Achim Hoffmann
School of Computer Science and Engineering, University of New South Wales, Sydney 2052, Australia
Keywords: Ontology Population, Semantic Role, Knowledge Acquisition.
Abstract: Ontology population is a knowledge acquisition activity that relies on (semi-) automatic methods to
transform unstructured, semi-structured and structured data sources into instance data. In this work, a
semantic-role based process for ontology population is presented that provides a suitable framework for
textual knowledge acquisition in the biological domain. In particular, with our approach, a given ontology
can be enriched by adding instances gathered from biological natural language texts. Our system’s modular
architecture provides a greater versatility than current approaches in the mentioned domain, as the process
of ontology population is not directly dependent on the linguistic rules developed from the corpus.
1 INTRODUCTION
The exponential growth of scientific literature in the
biomedical domain makes it difficult for researchers
to access the massive amounts of online information
and to keep abreast of biomedical knowledge that
spreads at an increasing rate.
In recent years, several techniques for
discovering, accessing, and sharing knowledge from
medical literature have been developed, including a
remarkable number of studies on discovering
various kinds of knowledge by mining the medical
literature (Friedman et al., 2006).
The Semantic Web is viewed as an extension of
the current Web, in which information is given well-
defined meaning, better enabling computers and
people to work in cooperation. The knowledge
representation technology used in the Semantic Web
is the ontology, which provides the meaning and
facilitates the search for contents and information
(Jiang et al., 2009). So, in complex domains such as
biomedicine, ontologies are used for organizing and
sharing biological knowledge as well as integrating
different sources of knowledge in order to provide
interoperability among different research
communities.
Ontologies have captured the interest of the entire
biomedical community (Rubin et al., 2007), in part
due to impact and success of the Genome Ontology
(GO) (Lewis, 2005). But the manual building of
ontologies is a tedious, time consuming task which
results in a knowledge acquisition bottleneck. So,
automated ontology learning methods have been
proposed to allow a reduction in the time and effort
needed in the ontology development process
(Valencia-García et al., 2008).
Ontology Learning (also named ontology
generation or ontology extraction) is a knowledge
acquisition activity that relies on (semi-) automatic
methods to transform unstructured (e.g. corpora),
semi-structured (e.g. folksonomies, html pages, etc.)
and structured data sources (e.g. databases) into
conceptual structures. Some ontology learning
approaches, such as TERMINAE (Aussenac-Gilles
et al., 2008), provide guidance to conceptualization
from natural language text integrating functions for
linguistic analysis and conceptual modelling.
Ontology Population, on the other hand, is a
knowledge acquisition activity that relies on (semi-)
automatic methods to transform unstructured, semi-
structured and structured data sources into instance
data. Thus, while Ontology Learning deals with the
acquisition of new concepts and relations with the
27
Maria Ruiz-Martinez J., Valencia-García R., Martínez-Béjar R. and Hoffmann A..
POPULATING BIOMEDICAL ONTOLOGIES FROM NATURAL LANGUAGE TEXTS.
DOI: 10.5220/0003065000270036
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2010), pages 27-36
ISBN: 978-989-8425-29-4
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)

consequence of changing the definition of the
ontology itself, the goal of Ontology Population is
the extraction and classification of instances of the
concepts and relationships defined in the ontology.
The instantiation of the ontology with new
knowledge is a relevant step towards the provision
of valuable ontology-based knowledge services.
In this work, a scalable methodology for
ontology population from textual resources based on
Natural Language Processing (NLP) and ontological
engineering techniques is proposed. This
methodology attempts to support expert
communities in building ontologies from natural
language texts. It allows several semantic relations
to be used and reduces the degree of expert
participation during the ontology construction
process. Our methodology has been implemented in
the form of a software prototype and tested in the
biomedical domain.
The structure of the paper can be described as
follows. In Section 2 related works are shown,
whereas Section 3 presents the Technical
Background. In Section 4, the Ontology population
process is described. Finally, some conclusions are
put forward in Section 5.
2 RELATED WORK
During the last decade several approaches that
(semi-)automatically build domain-specific
ontologies have been proposed, some of them just
generating hierarchies (taxonomies) of concepts,
(Cimiano et al., 2005) or relating concepts with a
reduced set of semantic relationships (Maedche et
al., 2001) (He, 2006). In biomedicine, taxonomy and
partonomy relationships are an important starting
point, although they are not enough for modelling
such a complex domain.
Some ontology learning methods enrich pre-
existing ontologies. For example, in (Agirre et al.,
2000) a methodology for enriching the concepts of
WordNet is presented. In (Sánchez and Moreno,
2008), a domain ontology is enriched by discovering
non-taxonomic relationships from the web using
patterns based on verb phrases. Other works such as
(Bada et al., 2007) focus on the enrichment of
biological ontologies through integration processes.
Regarding the biological domain, a number of
biological NLP and text mining systems have been
developed to extracting biological information and
knowledge. Most of them include a module that
recognizes biological entities or concepts in text,
usually called Named Entity Recognition (NER).
Biological NER is the task of identifying the
boundary of a substring and then mapping it onto a
predefined category (e.g., Protein, Gene or Disease).
Once the biological terms have been extracted, the
semantic relations can then be detected. In
(Bundschus et al., 2008), semantic relations between
diseases and treatments are classified using
Conditional Random Fields. In (Chun et al., 2006),
relationships between genes and diseases from
MedLine abstracts are obtained by studying the co-
occurrence of terms.
Other approaches (He, 2006) use machine
learning techniques and discourse analysis methods
in order to extract protein to protein interactions,
however they just considered taxonomy and
partonomy relationships.
In (Rosario and Hearst, 2004), the authors
identify semantic relations between “treatment” and
“disease” in bioscience texts by means of graphical
models and a neural network. Semantic role
labelling approaches have been developed to extract
the semantic relations. For example, PASbio
(Wattarujeekrit et al., 2004) is an extended model of
PropBank applied to Molecular Biology. The work
introduces the notion of semantic analysis of
argument roles in biological texts and proposes the
construction of Predicate Argument Structures
(PAS) for Molecular Biology. PAS are knowledge
structures that represent the relations between a verb
and its arguments. These predicates describe the
roles of genes and gene products in mediating their
biological functions. BioFrameNet (Dolbey et al.,
2006) is an extension of FrameNet that has added
semantic frames relevant to the Molecular Biology
domain. The semantics have been implemented in
OWL DL to facilitate links to domain ontologies
like GO or EntrezGene. Finally, BIOSMILE (Tsai et
al., 2007) is another semantic role labelling system
that was trained on BioPop, a biomedical
proposition bank semi-automatically annotated
consisting of 30 biomedical verbs that were
annotated into 500 abstracts. This system
incorporates lemmatized forms together with Part-
of-Speech tags and NE types.
3 TECHNICAL BACKGROUND
3.1 Ontologies
In this work, an ontology is seen as “a formal and
explicit specification of a shared conceptualisation”
(Studer et al., 1998). Ontologies provide a formal
and structured knowledge representation that has the
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
28

advantage of being reusable and shareable. In our
methodology, ontologies are obtained as a result of
knowledge extraction processes.
The Web Ontology Language (OWL) has been
used to represent the biological ontologies that are to
be populated from texts. In OWL, the main
ontological entities are classes, “subclass of”
relationships, datatype properties, object properties,
and individuals. OWL provides a formal theory for
taxonomy, whereas any other semantic relationship,
like partonomy or topology, must be manually
defined and implemented using object properties for
that.
In (Smith et al., 2005), the most common
relations used in biomedical domain ontologies were
presented and formalized. As a result of this effort,
the OBO ontology of biomedical relations was
produced. The OBO Relation Ontology
(http://www.obofoundry.org/ro) comprises ten
different types of relations including taxonomic and
partonomic relations. In this work, an ontological
model based on the different types of relations
defined in OBO Relations Ontology, has been
defined. This ontological model has been
implemented using the new version of the OWL
language, namely OWL 2, that adds several new
features to OWL, including increased expressive
power for properties and extended support for
datatypes.
Table 1: OWL 2 Property axioms of the semantic relations
in OBO relation ontology.
Relation T S R I A F IF
is a X X X
part_of X X X
located_in X X
contained_in X
adjacent_to
transformation_of X
derives_from X
preceded_by X
has_participant
has_agent
instance of
These relations are binary, and they have been
implemented using object properties and the
property axioms that can be defined in OWL 2 as
shown in Table 1. The main OWL 2 property
axioms are described in the following:
Reflexive (R). (X Relation X)
Irreflexive (I). not(X Relation X).
Symmetric (S). (X Relation Y) ↔ (Y Relation
X)
Asymmetric (A). (X Relation Y) →not(Y
Relation X).
Transitive (T). (X Relation Y) and (Y Relation
Z) → (X Relation Z).
Functional (F). (X Relation Y) and (X Relation
Z) → (Y = Z)
Inverse Functional (IF). (X Relation Y) and (Z
Relation Y) → (X = Z)
An OWL formal model allows for performing
automatically a set of Description Logic inference
services, which can be supported by DL reasoners
(e.g., HermiT, Pellet2, Fact++, Racer) (Sirin and
Parsia, 2004):
Consistency checking, which ensures that an
ontology does not contain any contradictory
facts.
Concept satisfiability, which checks whether it is
possible for a class to have any instances. If a
class is unsatisfiable, then defining an instance of
the class will cause the whole ontology to be
inconsistent.
Classification, which computes the subclass
relations between every named class to create the
complete class hierarchy. The class hierarchy can
be used to answer queries such as getting all or
only the direct sub-classes of a class.
Realization, which finds the most specific classes
that an individual belongs to; or in other words, it
computes the direct types for each of the
individuals.
An OWL ontology can be viewed from a logical
point of view as a collection of axioms that must be
satisfied. This does not only include classes and
properties, but also restrictions such as disjoint
classes. Consistency is a critical issue in Ontology
Engineering. We say that an ontology is internally
inconsistent when some parts of it are inconsistent
with other parts of itself. For instance, an ontology is
internally inconsistent if one of the properties
concerning relationships between concepts is not
satisfied. The property axioms defined in Table 1
help to detect any inconsistency in the populated
ontology. For example, the part-of relation holds
both transitive and asymmetric properties, so it is not
possible to have a cycle inside a conceptual
partonomy. This ensures the correct results of the
knowledge that can be inferred from the ontology by
applying the corresponding axioms. Moreover, the
POPULATING BIOMEDICAL ONTOLOGIES FROM NATURAL LANGUAGE TEXTS
29

existence of such restrictions is useful to grant the
consistency of the individuals built, which must
satisfy the restrictions defined for their
corresponding class. Moreover, the collection of
conditions defined for the classes can be used by the
reasoner for the automatic classification of
individuals.
3.2 Named Entity Recognition (NER)
The gap between linguistic biomedical texts and the
extraction and organization of their knowledge in
ontologies has been addressed primarily from the
extraction of terms and relations between them.
Terms extraction is a prerequisite for all aspects
of ontology learning from text. Terms are linguistic
realizations of domain-specific concepts and are
therefore central to further, more complex tasks.
(Buitelaar et al., 2005). In relation to current systems
and frameworks related to this work, UMLS stands
out as it merges information from more than 100
biomedical vocabularies, which makes existing
terminologies both easier to use and more useful
(Ananiadou and McNaught, 2006). However, not all
these terms will be names of biomedical entities
which are essential for populating the ontology.
In biomedical literature, NER refers to the task
of recognizing entity-denoting expressions such as
genes, proteins, cells and diseases (Ananiadou and
Mc Naught, 2006).
Ontology population systems share a general
architecture that is described in (Petasis et al., 2007)
and that consists of an extraction toolkit identifying
terms or NER in order to locate instances of
concepts and instances of relations between
concepts.
The majority of systems are rule and machine
learning based approaches (Saquete et al. 2008).
Fukuda (Fukuda et al., 1998) developed one of the
earliest NER systems for proteins, but the rules had
to be defined manually. In order to overcome these
problems, machine learning techniques have been
proposed. Some of the techniques are statistically
based (e.g., Hidden Markov models, Conditional
Random Fields, etc.). The advantage of machine
learning techniques is that they can identify potential
biomedical entities which are not previously
included in standard vocabularies.
In line with these techniques, the work presented
in (Settles, 2004) extracts Named Entities using
Conditional Random Fields. This method takes a set
of orthographic and semantic features into account
to train the system. Other works, such as (Shen et
al.,2003), use Hidden Markov models for NER in
the biological domain. Other machine learning
approaches are based on Support Vector Machines
(Lee et al., 2004). NER in our system is performed
by the GENIA Named Entities module (Kulick et
al., 2004), based on the GENIA corpus, combined
with grammatical patterns.
3.3 Semantic Role Labelling
A semantic role is the relationship between a
syntactic constituent and a predicate. It defines the
role of a verbal argument in the event expressed by
the verb (Moreda et al, 2010).
The semantic roles set developed in the
Proposition Bank (PropBank) project (Palmer et al.,
2005) and in the FrameNet project (Filmore, 2002)
are the most widely used in the literature.
In the biological domain, the most important sets
of semantic roles are PASbio (Wattarujeekrit et al.,
2004), BioFrameNet (Dolbey et al., 2006) and
BIOSMILE (Tsai et al., 2007) and they have been
used for extracting semantic relations and named
entities in biological domains.
In this work, the semantic roles provided by
PASbio are used for extracting and detecting
ontological relationships (amongst those defined in
Table 1) between the named entities extracted in
order to obtain and insert the individuals of the
domain ontology. In Figure 1, an example of a part
of the transform frame in PASbio is shown. This
pattern models the relation between an entity that is
going to be transformed into a new state by an agent
through the verb transform.
<predicate lemma="transform">
<roleset id="transform.01">
<roles>
<role n="0" descr="agent of transformation"/>
<role n="1" descr="entity undergoing transformation"/>
<role n="2" descr="end state"/>
</roles>
</roleset>
For expression of the recombinant protein,
[
1
pET28a-5] was transformed into
[
2
Escherichia coli strain BL21(DE3)].
end state
entity
Figure 1: An example of the transform frame in PASbio.
In order to detect ontological semantic relations
between entities, a mapping between semantic
relations and the semantic roles has been done. For
example the PASbio transform frame shown in
Figure 1 has been associated with the ontological
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
30
relationship transformation_of in the following
manner:
transformation.01(role
0
, role
1
, role
2
) => role
2
transformation_of role
1
That is, in this frame there exists an ontological
relation transformation_of between the role
2
and
role
1
. For example, in the sentence shown in Figure
1 the relation [Escherichia coli strain BL21(DE3)]
transformation_of [pET28a-5], would be obtained.
A mapping between verbal expressions and
PASbio frames is also necessary. In table 2 an
excerpt of these mappings are showed.
Table 2: An excerpt of the mapping between verb
expressions, PASbio frames and the ontological
relationships.
Verb
expression
PASbio
Frame
Ontological
relationship
be transform
into
Transform Transformation_of
is altered by
was mutated
changes in
be susceptible
to modify
Modify Transformation_of
may develop Develop Derives_from/
transformation_of
be altered Alter Derives_from/
transformation_of
be generated
by/from
Generate Derives_from
is the result of Result Derives_from
resulting in
are
transcribed
from
Transcribe Derives_from
4 ONTOLOGY POPULATION
PROCESS
The main aim of the process proposed here is to
populate biological domain ontologies from natural
language text using NLP and semantic technologies.
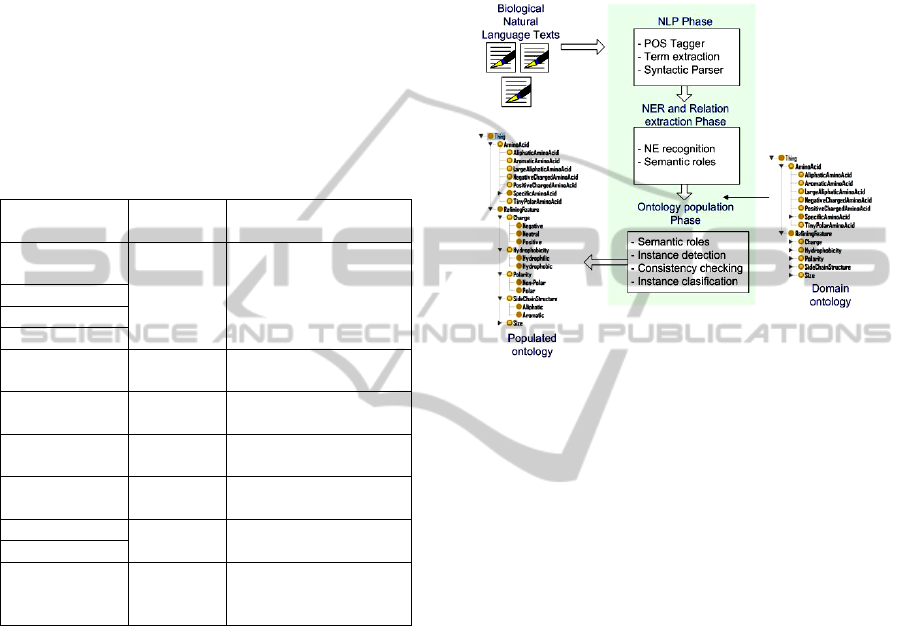
The architecture of the process is shown in Fig. 2. It
is composed of three main sequential phases: NLP
Phase, NER and relation extraction Phase and the
Ontology Population Phase. In a nutshell, the
process works as follows: in the NLP phase a lexical
and syntactical analysis of the corpus is done. Here
the grammar category of the words and the
constituents of each sentence are obtained. In the
second phase, the possible NEs and the semantic
relationships between them are extracted using NER
and semantic roles technologies. In the final phase,
the instances of the domain ontology are obtained
from the semantic annotations of the previous phase.
In this phase, the consistency of the instances and its
classification in the ontology are also addressed. An
enriched consistent domain ontology is obtained at
the end of this phase. In Figure 2, the referred phases
are shown.
Figure 2: Phases in the Ontology Population process.
4.1 NLP Phase
The main objective of this phase is to obtain the
morphologic and syntactic structure of each
sentence. For this, a set of NLP tools including a
sentence detection component, tokenizer, POS
taggers, lemmatizers and syntactic parsers has been
developed using the GATE framework
(http://gate.ac.uk/). GATE is an infrastructure for
developing and deploying software components that
process human language. GATE helps scientists and
developers in three ways: (i) by specifying an
architecture, or organizational structure, for
language processing software; (ii) by providing a
framework, or class library, that implements the
architecture and can be used to embed language
processing capabilities in diverse applications; (iii)
by providing a development environment built on
top of the framework made up of convenient
graphical tools for developing components.
Some aspects of biomedical texts may affect the
morphologic and syntactic analysis, such as the
ambiguity caused by names and abbreviations that
begin with capital letters; chemical and numeric
expressions including non-alphanumeric characters
such as commas, parentheses, and hyphens;
participles of unfamiliar verbs that describe domain-
specific events; and fragments of words (Tateisi and
Tsujii, 2004). The GENIA tagger (Tsuruoka et al.,
POPULATING BIOMEDICAL ONTOLOGIES FROM NATURAL LANGUAGE TEXTS
31

2005) is able to manage these problems more
efficiently than a general POS Tagger. Nevertheless,
not all the ambiguities are solved and, in some cases,
these unsolved issues can affect to later stages in the
process. Next, an example of a sentence analyzed
with GENIA is shown:
After/IN treatment/NN with/IN 5-
azacytidine/NN ,/, the/DT adult/JJ mesenchymal/JJ
stem/NN cells/NNS were/VBD transformed/VBN
into/IN cardiomyocytes/NNS ./.
Once the sentences have been analysed, a
lemmatization and the shallow parsing syntactic
analysis is done using Freeling (Atserias et al., 2006)
to obtain the main chunks of the sentence:
[After]
ptr
[treatment]
n
[with]
ptr
[5-azacytidine]
sn
[,]
sf
[the adult mesenchymal stem cell]
sn
[be
transform]
vb
[into]
prt
[cardiomyocytes]
n
.
4.2 NER and Relation Extraction
Phase
During this phase firstly the NE candidates are
identified by making use of the GATE Framework.
The output produced by each component of GATE
is a set of annotations, namely metadata associated
with a particular section of the document content.
Each annotation in the text is then merged into a
unified representation for each entity. All the
occurrences of identified NEs (NE mentions) in the
text will be candidate instances in the ontology. A
combination of JAPE rules and lists of Gazetters are
also used to perform the processes associated with
this phase.
Jape is a rich and flexible regular expression-
based rule mechanism offered by the GATE
framework (Sabou et al., 2005). On the other hand,
the gazetteer consists of a list of entities that are
relevant in the domain under question. Several lists
containing biological terms extracted from
GeneOntology (http://www.geneontology.org/) and
UMLS (http://www.nlm.nih.gov/research/umls/)
have been created. Examples of the general lists that
have been created are: Lipid, DNA, Aminoacid
monomer, Peptide, Organic compound, Multicellular
organisms, Cell type, etc. In Figure 3 an example of
the named entities recognized using GATE is
shown.
Once the NEs have been extracted, the PASbio
frames are detected in the text in order to extract the
possible relations between these named entities. For
example, in the previous example verb expression be
transform into is found, so the PASbio frame
transform has to be applied and that indicates that
there exists an ontological relationship
transformation_of between the named entities
obtained. That is cardiomyocytes transformation_of
mesenchymal stem cells.
Figure 3: Obtaining named entities with GATE.
4.3 Ontology Population Phase
Given a set of NEs and the ontological relationships
between them, during this phase the system firstly
determines if they are individuals of the ontology.
If so, the system must assign each individual to a
particular class of the domain ontology and inserted
it into the ontology. If some of the individual already
exist in the ontology then they are not inserted.
Each type of NE is associated with a Class of the
Ontology, so that, those NE mentions that in the
previous phase have been classified into a type of
NE become candidates for Ontology Instances.
These candidates are provisionally inserted in the
corresponding class or classes. Once the reasoner
checks the consistency of the ontology, those
instances that are inconsistent are deleted.
For example, Myosin heavy chain and troponin I
are NE mentions or occurrences of the type Protein.
The NE Protein is associated in the ontology with
the class Protein. So these NE mentions become
individuals of the Protein class. At the end of the
population process, the reasoner checks if the new
instances are consistent and if new knowledge can
be inferred.
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
32
Regarding the relationships between the individuals,
they are represented by means of object properties
between classes in the domain ontology. Due to the
fact that the domain ontology has been developed
using the ontological model described in section 3.1,
the object properties defined between the classes
have to be associated with any of the relationships
described in Table 1.
Once the individuals have been inserted into an
ontology, they have to be related using the
relationships identified in the previous phase. For
each of these relationships, the individuals that
participate in it are obtained from the ontology and
the system checks if they are already related by an
object property of the same type (i.e. part_of,
located_in, derives_from, etc). The relationship is
only inserted if the relationship does not exist in the
ontology, yet.
After that, a reasoner such as Pellet2 is executed
in order to (1) check for the consistency of the
ontology and (2) compute inferred types. If the
ontology is inconsistent, the last relationship inserted
into the ontology will be removed. In case that the
ontology is consistent, and the reasoner has inferred
that one individual belonging to the relationship can
be classified into a new class, this new classification
is done.
5 EVALUATION
A software prototype that implements this
methodology by means of a platform has been
developed for validating our approach. The platform
has been developed in Java, and the OWLAPI has
been used for processing the content of the
ontologies. The OWLAPI does not only provide a
rich API for dealing with OWL ontologies, but also
facilitates the use of OWL reasoners, so making it
possible to employ query languages such as
SPARQL. The NLP part is done using the GATE
framework along with some of the resources it
provides. A GENIA POS Tagger and Freeling plug-
in has been developed to integrate with GATE.
An ontology extracted from the xGENIA
ontology (Rak et al., 2007) has been used in the
experiment. The xGENIA ontology is an OWL-DL
ontology based on the GENIA taxonomy that was
developed as a result of manual annotation of the
GENIA corpus, which is a subset of the MEDLINE
one. Both the ontology and the corpus have been
used as a benchmark to test and develop biological
information extraction tools.
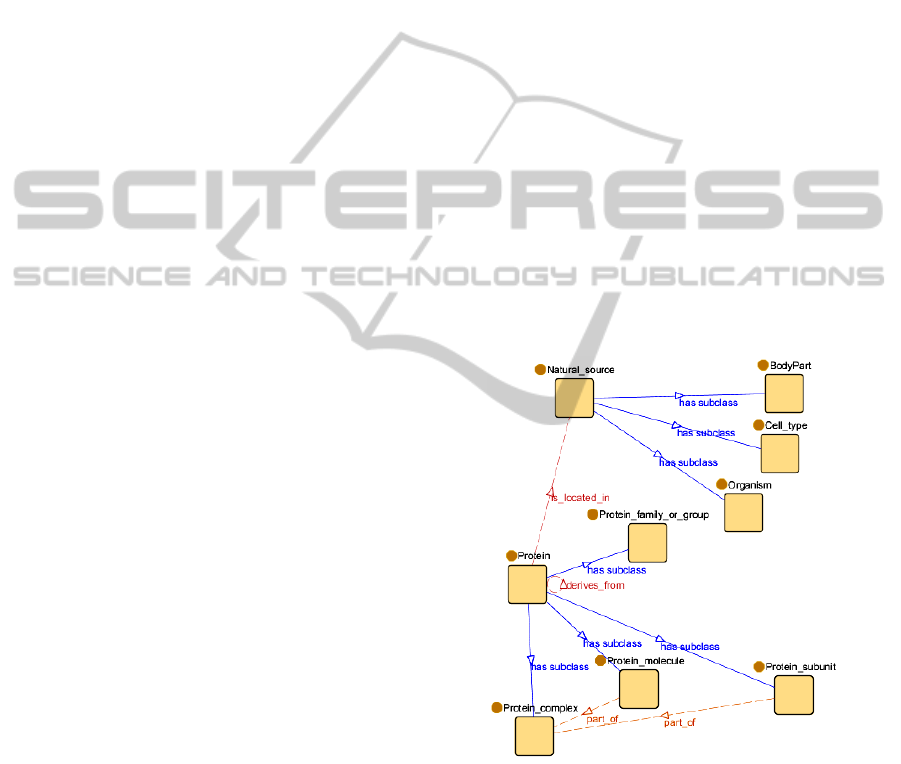
In Figure 4, the ontology that has been used for
validation is shown. The object properties between
classes in the xGENIA ontology are not based on the
most common relationships modelled by the OBO
relation ontology (see Table 1), so some changes in
these properties have been done in order to represent
them as a subset of the relations proposed in Table 1.
The ontology represents two hierarchies:
Protein: Proteins are organic compounds made
of aminoacids arranged in a linear chain and
folded into a globular form. Proteins include
protein groups, families, molecules,
complexes, and substructures. In the ontology
the following protein types have been
modelled: protein family or group, that is, a
family or a group of proteins; protein
complex, which includes conjugated proteins
such as lipoproteins and glycoproteins;
individual protein molecules, which are
individual members of a group of non-
complex proteins, and subunits of a protein
complex.
Natural source: Natural sources are biological
locations where substances are found and their
reactions take place. In this hierarchy only the
types, body part, cell type and organism have
been taken into account.
Figure 4: An excerpt of the ontology.
Apart from that, four non-taxonomic relations have
been defined:
Protein is_located_in Natural_source
Protein derives_from Protein
Protein_molecule part_of Protein_complex
Protein_subunit part_of Protein_complex
A small corpus extracted from the GENIA corpus
containing 3,798 words has been used for populating
POPULATING BIOMEDICAL ONTOLOGIES FROM NATURAL LANGUAGE TEXTS
33

and extracting the relations between the individuals
in the ontology.
Due to the fact that the prototype uses the
GENIA NER, the precision and recall of the NE
extraction is close to 96%. The aim of this
experiment was to evaluate the precision and recall
of the relation detection between instances in the
population process.
The resulting populated ontology has been
compared to the part of the xGENIA ontology which
is already instantiated.
Some frames representing part_of and
is_located_in relations have been manually
developed since PASbio frames are only based
onevent detection relationships in molecular
biology.
The precision score has been defined as the
number of ontological relationships extracted that
exist in the xGENIA ontology divided by the total
number of relationships extracted:
Another evaluation parameter used has been the
recall score, which has been defined as the number
of ontological relationships extracted divided by the
total number of ontological relationships that exist in
the corpus:
The prototype achieved a precision of 79.02% and a
recall of 65.4%. These values are significant because
(1) the domain is quite specific, and (2) the semantic
roles used have been designed for the biomolecular
domain.
6 CONCLUSIONS AND FUTURE
WORK
The semantic-role based process for ontology
population presented here provides a suitable
framework for textual knowledge acquisition in the
biological domain. In particular, with our approach,
a given ontology can be enriched by adding
instances gathered from biological natural language
texts.
In this work, the NER process is performed
using the GENIA Named Entities module, which is
based on machine learning techniques. Currently,
there exist many knowledge bases in the biomedical
domain such as UMLS and GeneOntology and the
use of such controlled vocabularies would be very
helpful for identifying NE and terms in biomedical
text. It is planned to develop a new NER module that
can also use these ontologies.
The performance of our system depends heavily
on the performance during the NER phase. Poor
perofrmance during this phase limits significantly
the system’s recall and precision.
On the other hand, the semantic relation
extraction is done using the reduced set of semantic
roles defined in PASbio. In order to improve the
semantic role detection, we are planning to include
more frames from BioFrameNet, BIOSMILE,
FrameNet and VerbNet to improve this issue.
Therefore, a set of predefined semantic relations
based on the OBO relation ontology have been
defined. However, some of the relationships
between the instances of the GENIA corpus cannot
be modelled with this set. BIOTOP (Beißwange et
al., 2008) is an upper domain ontology for biology
that adds some semantic relationships to the OBO
relation ontology such as, for example, has-
inherence, realization-of or has-grain relationships.
The system’s modular architecture gives our
system a greater flexibility, as the process of
ontology population is not directly dependent on the
linguistic rules developed from the corpus, such as
the approaches presented in (Tanev and Magnini,
2006) and (Amardeilh et al., 2005). The results of
the validation seem promising although they should
be compared with those of other ontology
construction methods. A more in-depth evaluation of
the system is planned, comprising the application of
the whole GENIA corpus and xGENIA ontology,
the use of statistical methods for analyzing the
results.and the comparison with some other
ontology population methods is also planned. The
validation of the proposed methodology within the
scope of other related domains such as the
biomedical domain is also left for future work.
ACKNOWLEDGEMENTS
This work has been possible thanks to the Spanish
Ministry for Education through grant JC2009-00194
under the program “José Castillejo”, and the
Regional Government of Murcia under project BIO-
TEC 06/01-0005. Juana María Ruiz-Martínez is
supported by the Fundación Séneca through grant
06857/FPI/07.
extractedipsrelationshtotal
extractedipsrelationshcorrect
precision =
corpusinipsrelationshtotal
extractedipsrelationshcorrect
recall =
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
34

REFERENCES
Amardeilh, F., Laublet, P., Minel, J.L. 2005, Document
annotation and ontology population from linguistic
extractions, Proceedings of the 3rd international
conference on Knowledge capture, , pp. 161-168.
Agirre, E., Ansa, O., Hovy, E. and Martinez, D. 2000,
Enriching very large ontologies using the WWW,
Proceedings of the ECAI Ontology Learning
Workshop in conjunction with the 14th European
Conference on Artificial Intelligence (ECAI 2000).
Ananiadou, S. & McNaught, J. 2006, Text mining for
biology and biomedicine, Artech House(ed).
Atserias, J., Casas, B., Comelles, E., González, M., Padró
L., and Padró, M (2006) FreeLing 1.3: Syntactic and
semantic services in an open-source NLP library.
Proceedings of the fifth international conference on
Language Resources and Evaluation (LREC 2006),
ELRA. Genoa, Italy.
Aussenac-Gilles, N., Despres, S., and Szulman., S, 2008
The TERMINAE Method and Platform for Ontology
Engineering from texts. Dans: Bridging the Gap
between Text and Knowledge - Selected Contributions
to Ontology Learning and Population from Text. Paul
Buitelaar, Philipp Cimiano (Eds.), IOS Press, p. 199-
223,
Bada, M. and Hunter, L. 2007, Enrichment of OBO
ontologies, Journal of Biomedical Informatics, vol. 40,
no. 3, pp. 300-315.
Beißwanger, E., Schulz, S., Stenzhorn H. and Hahn, U.
2008. BioTop: An Upper Domain Ontology for the
Life Sciences - A Description of its Current Structure,
Contents, and Interfaces to OBO Ontologies. Applied
Ontology, vol. 3, no. 4, pp. 205-212,
Buitelaar, P., Cimiano, P. and Magnini, B. 2005, Ontology
learning from text: An overview, Ontology learning
from text: Methods, evaluation and applications, , pp.
3-12.
Bundschus, M., Dejori, M., Stetter, M., Tresp, V. and
Kriegel, H. 2008, Extraction of semantic biomedical
relations from text using conditional random fields,
BMC Bioinformatics, vol. 9, no. 1, pp. 207.
Chun, H., Tsuruoka, Y., Kim, J., Shiba, R., Nagata, N.,
Hishiki, T. and Tsujii, J. 2006, Extraction of gene-
disease relations from MedLine using domain
dictionaries and machine learning, Pac Symp
Biocomput, vol. 11, pp. 4–15.
Cimiano, P., Pivk, A., Schmidt-Thieme, L. & Staab, S.
2005, Learning taxonomic relations from
heterogeneous sources of evidence, Proc of ECAI
2004 Workshop on Ontology Learning and
Population, pp. 59–73.
Dolbey, A., Ellsworth, M. and Scheffczyk, J. 2006,
BioFrameNet: A Domain-specific FrameNet
Extension with Links to Biomedical Ontologies,
Biomedical Ontology in Action KR-MED 2006
Proceedings, , pp. 87-94.
Filmore, C. 2002. Framenet and the linking between
semantic and syntactic relations. In Proceedings of the
19th international conference on computational
linguistics (COLING).
Friedman, C., Borlawsky, T., Shagina, L., Xing, H.R. and
Lussier, Y.A. 2006, Bio-Ontology and text: bridging
the modeling gap, Bioinformatics, vol. 22, no. 19, pp.
2421-2429.
Fukuda, K., Tamura, A., Tsunoda, T. & Takagi, T. 1998,
Toward information extraction: identifying protein
names from biological papers, Pacific Symposium on
Biocomputing, pp. 707-718.
He, X. 2006, A protocol for constructing a domain-
specific ontology for use in biomedical information
extraction using lexical-chaining analysis. Thesis
presented at University of Waterloo.
Jiang, X. and Tan, A. 2009, Learning and inferencing in
user ontology for personalized Semantic Web search,
Information Sciences, vol. 179, no. 16, pp. 2794-2808.
Kulick, S., Bies, A., Liberman, M. , Mandel, M.,
McDonald, R., Palmer, M., Schein A., and Ungar, L.
2004, Integrated Annotation for Biomedical
Information Extraction, HLT/NAACL 2004 Workshop:
Biolink, pp. 61-68.
Lee, K., Hwang, Y., Kim, S. and Rim, H. 2004,
"Biomedical named entity recognition using two-phase
model based on SVMs", Journal of Biomedical
Informatics, vol. 37, no. 6, pp. 436-447.
Lewis, S.E. 2005 Gene Ontology:looking backwards and
fowards, Genome Biol, vol.6, no.1, pp. 103.
Maedche, A. and Staab, S. 2001, Ontology Learning for
the Semantic Web, IEEE Intelligent Systems, vol. 16
(2), pp. 72-79.
Moreda P., Llorens H., Saquete E., and Palomar M., 2010
Combining semantic information in question
answering. Information Processing and Management.
Article in Press, Corrected Proof
Palmer, M., Gildea, D. and Kingsbury, P. 2005. The
proposition bank: An annotated corpus of semantic
roles. Computational Lingustics, no.31, vol.1, pp.71-
106.
Petasis, G., Karkaletsis, V. and Paliouras, G. 2007,
Ontology population and enrichment: State of the art.
Deriverable d4.3, BOEMIE: Bootstrapping Ontology
Evolution with Multimedia Information Extraction.
Rak, R., Kurgan, L., and Reformat, M. 2007, xGENIA: A
comprehensive OWL ontology based on the GENIA
corpus. Bioinformation.vol.1, no.9, pp.360–362.
Rosario, B. and Hearst, M.A. 2004, Classifying semantic
relations in bioscience texts, Proceedings of the 42nd
Annual Meeting on Association for Computational
Linguistics.
Rubin, D. L., Shah, N.H. and Noy, N.F. 2008, Biomedical
ontologies: a functional perspective, Briefings in
Bioinformatics, vol. 9, no. 1, pp. 75-90.
Sabou, M., Wroe, C., Goble, C. & Stuckenschmidt, H.
2005, Learning domain ontologies for semantic Web
service descriptions, Web Semantics: Science,
Services and Agents on the World Wide Web, vol. 3,
no. 4, pp. 340-365.
Sánchez, D. and Moreno, A. 2008, Learning non-
taxonomic relationships from web documents for
POPULATING BIOMEDICAL ONTOLOGIES FROM NATURAL LANGUAGE TEXTS
35

domain ontology construction, Data & Knowledge
Engineering, vol. 64, no. 3, pp. 600-623.
Saquete, E., Ferrández, O., Ferrández, S., Martínez-Barco,
P. & Muñoz, R. 2008, Combining automatic
acquisition of knowledge with machine learning
approaches for multilingual temporal recognition and
normalization, Information Sciences, vol. 178, no. 17,
pp. 3319-3332.
Settles, B. 2004, Biomedical Named Entity Recognition
Using Conditional Random Fields and Rich Feature
Sets, Proceedings of the International Joint Workshop
on Natural Language Processing in Biomedicine and
its Applications (NLPBA), vol. 1, pp. 104-107.
Shen, D., Zhang, J., Zhou, G., Su, J. & Tan, C.L. 2003,
Effective adaptation of a Hidden Markov Model-based
named entity recognizer for biomedical domain,
Proceedings of the ACL 2003 workshop on Natural
language processing in biomedicine, vol. 13, pp. 49-
56.
Sirin, E., Parsia, B. 2004. Pellet: An OWL DL reasoner.
Proc. of the 2004 Description Logic Workshop (DL
2004), pp. 212–213.
Smith B, Ceusters W, klagges B, Köhler J, Kumar A,
Lomax J, et al. 2005, Relations in biomedical
ontologies. Genome Biology,no.6, vol.5. R46.
Studer R, Benjamins VR, Fensel D., 1998, Knowledge
engineering: Principles and methods. Data
Knowl.Eng. no.25, vol.1-2 pp.161-197.
Tanev, H. & Magnini, B. 2006, Weakly Supervised
Approaches for Ontology Population, Proceedings of
EACL-2006, Trento, pp. 3-7.
Tateisi, Y. & Tsujii, J. 2004, Part-of-Speech Annotation of
Biology Research Abstracts, Proceedings of LREC04.
Tsai, R., Chou, W., Su, Y., Lin, Y., Sung, C., Dai, H.,
Yeh, I., Ku, W., Sung, T. and Hsu, W. 2007,
BIOSMILE: A semantic role labeling system for
biomedical verbs using a maximum-entropy model
with automatically generated template features, BMC
Bioinformatics, vol. 8, no. 1, pp. 325.
Tsuruoka, Y, Tateishi, Y, Kim, J, Ohta, T, McNaught, J,
Ananiadou, S, and Tsujii, J., 2005 Developing a
Robust Part-of-Speech Tagger for Biomedical Text,
Advances in Informatics - 10th Panhellenic
Conference on Informatics, LNCS vol.3746, pp. 382-
392.
Valencia-García,R. Fernández-Breis, J.T., Ruiz-Martínez,
J.M., García-Sánchez, F. and Martínez-Béjar, R. A
knowledge acquisition methodology to ontology
construction for information retrieval from medical
documents.2008, Expert Systems: The Knowledge
Engineering Journal vol.25, no.3, pp. 314-334.
Wattarujeekrit, T., Shah, P. and Collier, N. 2004, PASBio:
predicate-argument structures for event extraction in
molecular biology, BMC Bioinformatics, vol. 5, no. 1,
pp. 155.
KEOD 2010 - International Conference on Knowledge Engineering and Ontology Development
36
