
INVESTIGATING REPLACEMENT STRATEGIES FOR THE
ADAPTIVE DISSORTATIVE MATING GENETIC ALGORITHM
Carlos M. Fernandes, Juan Julián Merelo
Department of Computers’ Architecture, University of Granada, Granada, Spain
Agostinho C. Rosa
Department of Electrotechnics, Technical University of Lisbon, Lisbon, Portugal
Keywords: Genetic algorithms, Dissortative mating, Dynamic optimization problems.
Abstract: This paper investigates the effects of modifying the Adaptive Dissortative Mating Genetic Algorithm
(ADMGA) replacement strategy on the performance of the algorithm in dynamic problems. ADMGA is a
variation of the standard GA with a mating restriction based on the genotypic similarity of the individuals.
Dissimilar individuals mate more often than expected by chance and, as a result, genetic diversity
throughout the run is maintained at a higher level. ADMGA was previously tested in dynamic optimization
problems with promising results: the algorithm shows to outperform standard GAs and state-of-the-art
approaches on several problems and dynamics. However, the performance of the algorithm degrades when
the frequency of changes increases. Due to the premises under which ADMGA was tested, it has been
argued that the replacement strategy that emerges from the algorithm’s dissortative mating strategy may be
harming the performance in such situations. This study proposes alternative replacement schemes with the
objective of improving ADMGA’s performance on fast changing environments (without damaging the
performance on slower ones). The strategies maintain the simplicity of the algorithm, i.e., the parameter set
is not increased. The replacement schemes were tested in dynamic environments based on stationary
functions with different characteristics, showing to improve standard ADMGA’s performance in fast
dynamic problems.
1 INTRODUCTION
In the last two decades, Evolutionary Algorithms
have been successfully applied in industrial
problems, especially those with non-linearities and
multiple objectives. However, real-world problems
often have dynamic components that lead to
(predictable or unpredictable) variations of the
fitness function, i.e., the problem is defined by a
time-varying fitness function.
A problem is said to be dynamic when there is a
change in the fitness function, problem instance or
restrictions, thus making the optimum change as
well. In each period of optimization, the fitness
function is deterministic, but when changes occur,
solutions already found may be no longer valid and
the process must engage in a new search effort.
Evolutionary Algorithms’ (EAs) self-adaptive
characteristics make them promising candidates to
solve this type of problems.
Nowadays, these research efforts on evolutionary
dynamic optimization are being mainly directed
towards diversity maintenance techniques and
memory schemes. There are other possible
approaches, like reacting to changes (Cobb, 1990)
when they occur, or using multi-populations (Branke
et al., 2000), or even tackle the change with a new
randomly generated population, but the performance
of such kind of approaches is strongly dependent on
the intensity of the changes — they perform better
when changes affect a small percentage of the
genotype’s variables — and, usually, require that the
changes are easy to detect. Moreover, even if the
change is easy to detect, it is hard to decide whether
it is better to restart the population or continue the
search with the same population after a shift in the
environment. Thus, it is sometimes better to have an
algorithm that is capable of continuously adapting
104
Fernandes C., Merelo J. and Rosa A..
INVESTIGATING REPLACEMENT STRATEGIES FOR THE ADAPTIVE DISSORTATIVE MATING GENETIC ALGORITHM.
DOI: 10.5220/0003087001040113
In Proceedings of the International Conference on Evolutionary Computation (ICEC-2010), pages 104-113
ISBN: 978-989-8425-31-7
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)

the solution to a changing environment — for
instance, by reusing the information gained in the
past via a memory. For a description of a possible
set of categories to classify evolutionary approaches
to dynamic problems, please refer to (Branke, 2001).
Memory schemes (Goldberg & Smith, 1987;
Ramsey & Grefenstette, 1998) may be very effective
in some situations, and overcome some of the
aforementioned difficulties but their utility is
believed to be restricted to a certain type of
dynamics — in general, memory is particularly
useful when the dynamics of change is circular, i.e.,
the shape of the fitness landscape repeats from time
to time. In addition, they require a considerable
tuning effort and some parts of their design and
implementation may be not trivial (Branke, 1999).
Diversity maintenance techniques (Grefenstette,
1992; Yang, 2008; Tinós & Yang, 2007; Fernandes
et al., 2008c) usually slow down the convergence of
the algorithm during the stationary periods, a
characteristic that may harm the performance when
the changes in the fitness function are separated by
short periods of time (high frequency of changes).
However, these approaches do not require, in
general, any knowledge about the problem and
neither its dynamics nor its performance is reported
to be highly dependent on a specific configuration of
the problem.
A possible approach for designing diversity
maintenance EAs for dynamic optimization is using
mating restrictions based on the genotypes.
Dissortative mating, for instance, which refers to
mating strategies in which dissimilar individuals
mate more often than expected by chance, may be
inserted into to an EA and slow down the diversity
loss. There are several EAs in the literature with
such type of mating strategies. One of them is the
Adaptive Dissortative Mating Genetic Algorithm
(ADMGA), proposed by Fernandes and Rosa
(2008a) and applied to dynamic optimization with
promising results in (Fernandes & Rosa, 2008b;
Fernandes, 2009). However, it has been observed
that its performance degrades when the frequency of
changes increases. One of the possible explanations
for this behavior resides in the replacement strategy
and the premises under which it is tested: since
changes are assumed to be hard to detect, the
algorithm reevaluates every solution that remains in
the population after one generation (Please note that
this is the worst case scenario; in many applications
the changes may be detected with less computational
effort).
This problem arises because ADMGA’s
population replacement procedure is a population-
wide elitist strategy (Thierens, 1999): parents and
children compete and live in the same population. It
has been shown (Fernandes, 2009) that if every old
solution is reevaluated, then the average ratio
between ADMGA’s new individuals and function
evaluations, in each generation, is approximately ½.
In addition, since the replacement strategy is elitist,
it tends to reduce diversity. This may be slowing
down ADMGA and the effect is much more
pronounced with high frequency of changes because
there are fewer evaluations them.
This paper addresses this issue by proposing
alternative replacement strategies that introduce
diversity in the ADMGA’s parents’ subpopulation.
Three different schemes are proposed: one in which
the parents that remain in the population are first
mutated and then reevaluated: Replacement Strategy
2 (RS 2); another one that replaces the parents by
mutated copies of the best individuals (RS 3);
finally, a third scheme that is inspired by the
Random Immigrants Genetic Algorithm (RIGA)
(Grefenstette, 1992) and replaces the parents that
remain in the population by randomly generated
solutions (RS 4). The three strategies are tested in
several dynamic problems designed with a dynamic
problem generator (Yang, 2003). The results are
compared to those attained by the standard
ADMGA, here also described as Replacement
Strategy 1 (RS 1). Then, the best strategy is
compared with a standard Generational Genetic
Algorithm (GGA) and with a recently proposed
evolutionary approach for dynamic optimization,
called Elitism-based Immigrants Genetic Algorithm
(EIGA) (Yang, 2008). The results demonstrate that
the best strategy (RS 2) is clearly capable of
outperforming standard ADMGA on fast
environments, without degrading its performance
when the frequency is lower. The new algorithm
increases the frequency value below which ADMGA
is better than or equivalent to GGA and EIGA.
Statistical tests are provided.
The paper is structured as follows. The following
section briefly describes the most relevant
dissortative mating strategies found in literature.
Section 3 describes ADMGA and introduces the new
replacement strategies. Section 4 describes the
experimental setup and Section 5 presents and
discusses the results. Finally, Section 6 concludes
the paper and outlines future lines of research.
2 PREVIOUS WORK
By considering merely the quality of the solutions
INVESTIGATING REPLACEMENT STRATEGIES FOR THE ADAPTIVE DISSORTATIVE MATING GENETIC
ALGORITHM
105

represented by the chromosomes when selecting
individuals for mating purposes, the traditional GAs
emulate what, in nature, is called random mating
(Russel, 1998), i.e., mating chance is independent of
genotypic or phenotypic distance between
individuals. However, random mating is not the sole
mechanism of sexual reproduction observed in
nature. Outbreeding, assortative mating and
dissortative mating (Russel, 1998) are all non-
random strategies frequently found in the behavior
of natural species. These schemes have different
effects on the genetic diversity of the population.
Take for instance dissortative mating, which is
known to increase the diversity of a population
(Russel, 1998). Assortative mating, on the other
hand, restricts mating between dissimilar individuals
and leads to diversity loss.
Therefore, dissortartive mating naturally came
out in EAs’ research field as an inspiration for
dealing with the problem of genetic diversity and
premature convergence. A well-known GA with a
dissortative mating strategy is the CHC (Eschelman,
1991). CHC uses no mutation in the classical sense
of the concept, but instead it increases the mutation
probability when the best fitness does not change
after a certain number of generations. A
reproduction restriction assures that selected pairs of
chromosomes will reproduce unless their Hamming
Distance is above a certain threshold, that is, the
algorithm restricts crossover between similar
individuals. Another possible way of inserting
assortative or dissortative mating into a GA is
described in (Fernandes & Rosa, 2001). The
negative Assortative Mating GA (nAMGA) selects,
in each recombination event, one parent, by any
method. Then, it selects a pool of individuals —
the size of the pool controls the intensity of mating
restriction — and computes the Hamming distance
between those chromosomes and the first parent.
The individual less similar to the first parent is
selected for recombination. Although nAMGA’s
results are interesting, the size of the pool is critical
to its performance and hard to tune.
Ochoa et al. (2005) carried out an idea related
with nAMGA in a dynamic optimization framework.
Assortative and dissortative GAs are used to solve a
dynamic knapsack problem. The results show that
dissortative mating is more able to track solutions,
while a standard GA often fails to track them. The
assortative GA is the worst algorithm in the test set.
The authors also discuss the optimal mutation
probability for different strategies, concluding that
the optimal value increases when the strategy goes
from dissortative to assortative. In this line of work,
there is also a study by Ochoa et al. (2006) on the
error threshold of replication in GAs with different
mating strategies that aims at shedding some light
into the relationship between mutation probabilities
and mating strategies in EAs. The report reinforces
the idea that any experimental study on non-random
mating strategies for EAs must take into account
several mutation probability values; otherwise, the
results are probably biased towards a specific
strategy.
Besides the above-referred techniques, a large
number of other GAs with non-random mating are
found in the literature. Due to their characteristics,
these GAs are worthwhile exploring as diversity
maintenance schemes for dynamic optimization.
3 ADMGA AND REPLACEMENT
STRATEGIES
There are many possible replacement strategies
1
for
GAs but, in general, they may be classified into two
categories: generational and elitist. Generational
GAs replace the entire parents’ population by the
children; in elitist strategies, offspring has to
compete with their parents. ADMGA, due to its
specific design, is a population-wide elitist strategy
(Thierens, 1999). This means that some individuals
may remain in the population for more than one
generation. Since changes in non-stationary
functions are not always easy to detect, the most
reliable way to guarantee that a fitness value does
not become outdated by a change in the environment
is to reevaluate all the chromosomes that remain in
the population after reproduction. Assuming this
worst case scenario does not affect generational
GAs, because the entire population is replaced by
the offspring in each generation, and fitness values
must be always computed — where is the
population size —, independently of the premises.
As for an elitist GA, assuming that changes are
very hard to detect means that old individuals must
be reevaluated and that the average ratio between
new solutions and function evaluations, in each
generation, is below 1. In the particular case of
ADMGA, it has been shown (Fernandes, 2009) for
several problems that this ratio is approximately ½,
meaning that, ADMGA generates only half of the
solutions that a standard generational GA is able to
1
We call replacement strategy to the procedure that, from the
population of parents P(t) and the population of offspring P’(t),
selects the individuals that form the population P(t+1) and then
replace population P(t).
ICEC 2010 - International Conference on Evolutionary Computation
106

generate in the same period of time. This may be
particularly penalizing when the frequency of
changes is high, and, in fact, ADMGA’s
performance has been shown to degrade in those
situations. The question is: is it possible to improve
ADMGA’s performance in fast dynamic problems
by changing the replacement strategy in a way that
those reevaluations are accompanied by the
introduction of new genetic material in the
population? To assess this hypothesis, we test three
alternative replacement strategies. Before discussing
them, let us describe the main algorithm.
ADMGA is a self-regulated dissortative mating
EA, which incorporates an adaptive Hamming
distance mating restriction that tends to relax as the
search process advances. After two parents are
selected, crossover only occurs if the Hamming
distance between them is found to be above a
threshold value. If not, the recombination event is
classified as failed and another pair of individuals is
selected until 2
⁄
pair have tried to recombine ( is
the population size).
ADMGA
initialize population P(t) with size N
evaluate population P(t)
set initial threshold ts(0)
while (not termination condition)
create new individuals P’(t)
evaluate new individuals P’ (t)
create new population // see figure 2
end while
Procedure: create new individuals
matingEvents ← /2;
successfulMating ← 0;
failedMating ← 0
while (successfulMatings < 1) do
for (i ← 1 to matingEvents) do
select two chromosomes (c
1
, c
2
)
compute Hamming distance H(c
1
, c
2
)
if (H(c
1
, c
2
) >= ts(t))
crossover and mutate
successfulMating ← successfulMating+1
end if
if (H(c
1
, c
2
) < ts(t)) failedMating
←failedlMating+1
end for
if (failedMating > successfulMating) ts(t+1)← ts(t)-
1
else ts(t+1) ← ts(t)+1
end while
Figure 1: ADMGA’s pseudo-code.
After the reproduction cycle is completed, a new
population is created by selecting the members
amongst the parents and newly generated offspring.
Then, the threshold is incremented when the number
of successful matings is greater or equal than the
number of failed matings, and it is decremented
otherwise (see pseudo-code in figure 1). This way,
the genetic diversity indirectly controls the threshold
value. When diversity is decreased, threshold tends
to be decremented because the frequency of
unsuccessful mating will necessarily increase.
However, mutation introduces variability in the
population, resulting in occasional increments of the
threshold that moves it away from 0. The only
parameters that need to be tuned in ADMGA is
population size and mutation probability
.
Crossover probability is not used (in a way,
is
somewhat adaptive, because selected individuals
recombine or not depending on their Hamming
distance and the threshold value). As for the
threshold, ADMGA has shown to be capable of self-
adapting its value in the first generation, and
therefore threshold may be set to its highest possible
value (1, where is the chromosome length) in
the beginning of the run. However, in order to avoid
initial generations in which the ratio between new
individuals and function evaluations is very low, an
initial threshold value of 4
⁄
is used when
optimizing non-stationary functions.
RS 1
insert best ’ individuals from P(t) into P’(t)
P(t+1) ← P’(t)´
// is the size o P(t) and ’ is the size of P’(t)
RS 2
insert mutated best ’ individuals from P(t) into
P’(t)
P(t+1) ← P’(t)
RS 3
insert ’ copies of mutated best from P(t) into
P’(t)
P(t+1) ← P’(t)
RS 4
insert ’ random solutions into P’(t)
P(t+1) ← P’(t)
Figure 2: ADMGA’s create new population procedure:
replacement strategies (RS).
DMGA was tested in dynamic optimization
problems and it showed to outperform a standard
generationl GA, a standard population-wide elitist
GA, RIGA, EIGA and the Self-Organized Criticality
RIGA (SORIGA) (Tinós & Yang, 2007) on several
problems and dynamics (Fernandes, 2009).
However, when the frequency of changes is high,
ADGMA’s performance when compared to the other
INVESTIGATING REPLACEMENT STRATEGIES FOR THE ADAPTIVE DISSORTATIVE MATING GENETIC
ALGORITHM
107

algorithms diminishes. In order to overcome this
difficulty, three different replacement strategies are
introduced. Figure 2 describes these replacement
strategies, as well as the original scheme used for
dynamic optimization (RS 1). Please note that every
strategy inserts the offspring into the new
population. The differences reside in the way in
which the remaining slots are occupied — that is,
slots, where is the population size and ’ is
the offspring population size.
Replacement strategy 1 (RS 1) — original
ADMGA’s strategy — inserts the best
individuals from the parents’ population into the
new population. Replacement strategy 2 (RS 2) fills
up the remaining slots with mutated copies of the
’ best individuals in parents’ population (with
mutation probability
). Replacement strategy 3
(RS 3) inserts ’ mutated copies of the best
solution. Finally, strategy 4 (RS 4) inserts random
immigrants — i.e., randomly generated genotypes
— into the vacant slots. The following section
describes the problems used to test the efficiency of
the algorithms.
4 EXPERIMENTAL SETUP
The experiments were conducted with dynamic
versions of an order-3 trap function, an onemax
problem and the 01 knapsack problem. This way
we have, in the test set, a simple linear function
(onemax), a quasi-deceptive trap function (order-3
trap) and a combinatorial problem (knapsack). The
stationary functions were then used to construct
dynamic versions via the dynamic problem
generator proposed in (Yang, 2003) This section
describes the stationary functions, the dynamic
problem generator, and the methodology followed
during the experiments.
The knapsack version used in these experiments
is described in (Yang & Yao, 2005). The function
has a global optimum with fitness 1853 (since the
weights are non-negative integers the global
optimum can be obtained with dynamic
programming). A trap function is a piecewise-linear
function defined on unitation (the number of ones in
a binary string) that has two distinct regions in the
search space, one leading to a global optimum and
the other leading to the local optimum. Depending
on its parameters, trap functions may be deceptive or
not. The traps in this study are defined by:
,
1
,
(1)
where u() is the unitation function and is the
problem size (and also the fitness of the global
optimum). With this equation, order-
traps are in
the region between deceptive and non-deceptive. For
this study, a 30 bit problem was constructed by
concatenating 10 order-3 subproblems. The fitness
of the global optimum is 30. Finally, the onemax is a
simple linear problem that consists in maximising
the number of ones in a binary string. For the
experiments, we used a 100-bit problem.
The test environment proposed in (Yang, 2003)
was then used to create a dynamic experimental
setup based on the functions described above. This
problem generator has two parameters that control
the severity of the changes and their frequency: is
a value between 0 and 1.0 which controls the
severity of change and defines number of
generations between changes. By changing and
it is possible to control two of the most important
features when testing algorithms on dynamic
optimization problems: severity () and period (
— i.e., 1
⁄
is the frequency — between changes
(Angeline, 1997). In order to evaluate an algorithm’s
configuration when solving a specific problem, the
offline performance (Tinós & Yang, 2007) — i.e.,
the best-of-generation fitness values averaged over
the total number of runs and over the data gathering
period — is first examined:
1
G
1
R
(2)
where is the number of generations, is the
number of runs (30 in all the experiments) and
is the best-of-generation fitness of generation of
run of an algorithm on a specific problem. This
value gives information on how close the GAs are
able to track the moving solution.
Problem generator’s parameter defines the
number of generations between each change.
Because this value, if provided without the
population size , does give us enough information
on the real period between changes, in this paper we
use the number of evaluations between each change
. This does not affect the generator because if
every individual in population (with size ) is
evaluated in each generation , then /.
For each one of the stationary problems, five
different dynamic scenarios were constructed by
setting to 600, 1200, 2400, 4800, 9600, 19200 and
38400. As for the severity () value, it is randomly
generated in each time the function changes. The
scope of this investigation is the performance
according to the frequency of changes, and therefore
ICEC 2010 - International Conference on Evolutionary Computation
108

setting to random values simplifies the analysis.
Every run covered 50 periods of change, i.e., 50
evaluations, with changes every evaluations.
A GA has several parameters that model their
general behavior. We are particularly interested in
GAs’ performance when varying the mutation
probability, because evolutionary approaches that
work by maintaining population diversity at a higher
level during the search may be shifting the optimal
mutation probability to different values. For
instance, and as stated above, it has been
demonstrated that dissortative and assortative mating
increase and decrease, respectively, the optimal
mutation probability of a GA. Therefore, it is of
extreme importance to test the GAs under a
reasonable range of
values, otherwise the results
may become biased toward some of the approaches.
Probability values
were set to 1/2, 1/, 2/
and 4/.
The population size also affects the
performance of the GAs, not only on static
problems, but also in dynamic environments.
Knowing the optimal size is important for
determining with accuracy the scalability of a GA
and to avoid superfluous computation effort due to a
population larger than the optimal. Although this
investigation does not aim at studying scalability or
finding the optimal population size for each
problem, a proper research method must test
different values, otherwise there is a risk of
comparing suboptimal parameter settings and,
consequently, getting invalid conclusions. In this
study, all the algorithms were tested with 8,
16, 30, 60 and 120.
As for crossover, uniform crossover was chosen
in order to avoid taking advantage of the trap
function building blocks tight linkage, which
happens when using other traditional operators such
as one- or two-point crossover. Every algorithm in
the test set uses binary tournament (tournament size
2 is in general a fairly good selective pressure for
most problem (Thierens, 1999)).
The ADMGA versions were compared with
GGA and EIGA. EIGA is a very simple scheme that
in each generation replaces a fraction
of the
population by mutated copies of the best solution of
the previous generation (with mutation
probability
). The author shows that the algorithm
is more effective when the changes are not too
severe. Due to its simplicity and the interesting
results reported in (Yang, 2008), EIGA was selected
as the main peer-algorithm for this study. In
addition, EIGA has some similarities with one of the
replacement strategies proposed in this paper to
improve ADMGA’s performance, which makes in a
suitable candidate for being included in the test set.
EIGA and ADMGA’s RS 4 are inspired by the
Random Immigrants GA (RIGA) (Grefenstette,
1992), which maintains diversity by introducing
random solutions in the population in each
generation, thus guarantying that brand new genetic
material enters the population in every time step.
Although RIGA is a kind of standard GA for
evolutionary dynamic optimization experiments, the
results in (Fernandes & Rosa, 2008b) and (Yang,
2008) show that ADMGA and EIGA are able to
clearly outperform RIGA in most of the dynamic
scenarios. Therefore, we chose to remove the
algorithm from the test set in order to simplify the
study and the report. Moreover, RS 4 was found to
be the worst replacement strategy in the test set,
being unable to deal with the proposed dynamic
problems. RS 4 is not a proper strategy for ADMGA
and therefore, in order to simplify the graphics, it
was removed from analysis and discussion in section
5.
GGA was tested with crossover probability set to
0.7 and 1.0. A 2-elitist GGA was also tested. The
best results were attained with
1.0 and 2-
elitism. Like the other algorithms, EIGA was also
tested with several
values;
is set to 0.6 (as
suggested in (Yang, 2008)), 0.7 and 1.0;
is set 0.2
(also, as suggested in (Yang, 2008)). Please note that
due to its design, EIGA population size
must set
so that
1
, where is the population
size of a standard GA that would perform the same
number of function evaluations in each generation.
EIGA was tested with different
values and the
results discussed in the following section refer
always to the best configurations. Please refer to
(Yang, 2008) for details on this particular issue and
on the algorithm’s implementation and parameter
tuning.
Figure 3: Experimental results with order-3 trap function.
Standard ADMGA compared with GGA and EIGA.
Population size 30;
1
⁄
(GGA) and
2
⁄
(EIGA and ADMGA);
1.0; GGA with 2-
elitism.
22
24
26
28
30
ε=1200 ε=2400 ε=4800 ε=9600 ε=19200
averagedoffline
performance
order‐3trap
ADMGA
GGA
EIGA
INVESTIGATING REPLACEMENT STRATEGIES FOR THE ADAPTIVE DISSORTATIVE MATING GENETIC
ALGORITHM
109
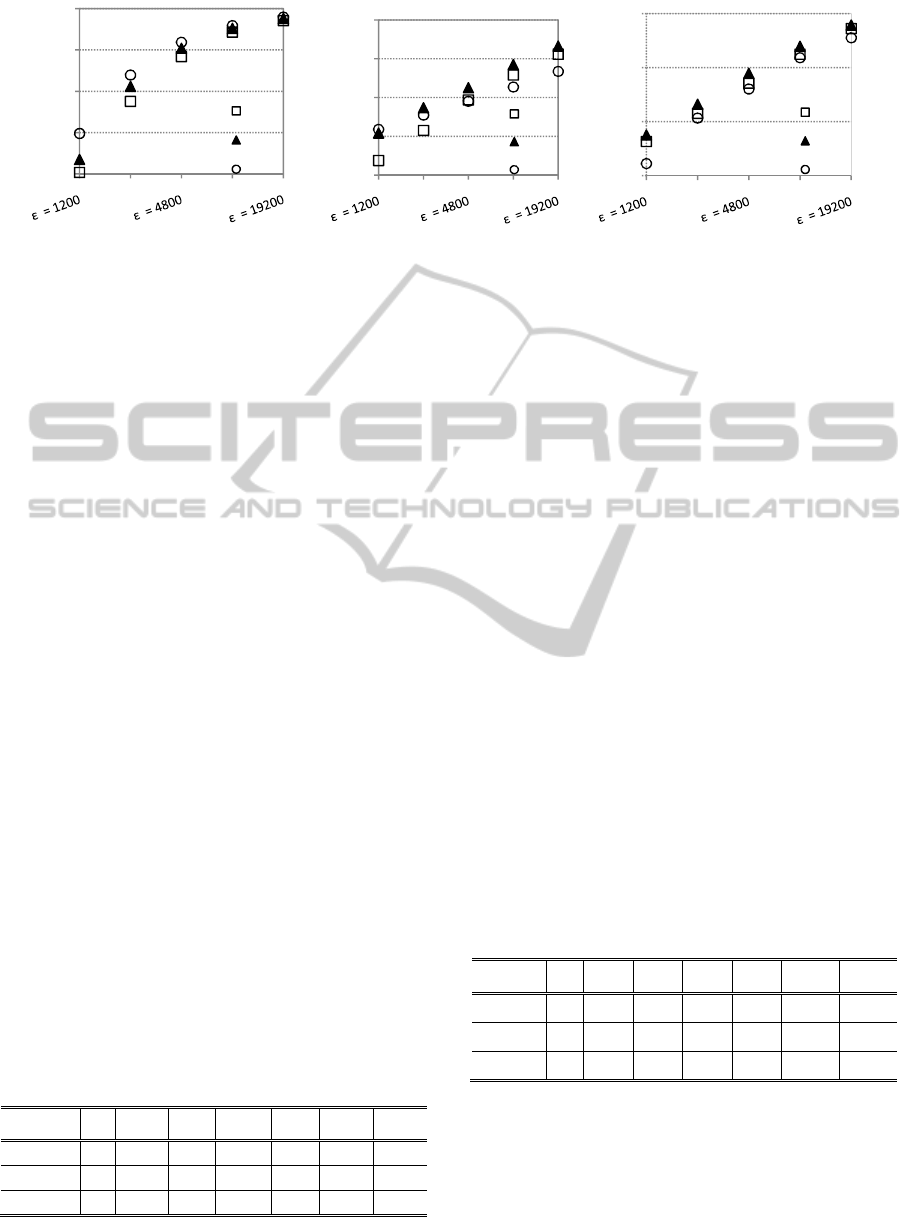
Figure 4: ADMGA replacement strategies in onemax, order-3 trap and knapsack dynamic problems. Population size:
16 (onemax) and 30 (trap and knapsack); 2/ (RS 1) and 1/ (RS 2 and RS 3). RS 2 with 2-
elitism.
5 RESULTS AND DISCUSSION
Figure 3 illustrates the issue addressed by this study.
ADMGA only outperforms the other GAs when is
above a specific value. In the order-3 dynamic
problem, ADMGA is clearly outperformed by the
other algorithms when 4800 — an assumption
confirmed by statistical tests. The main objective is
to find a replacement strategy for ADMGA that
reduces this value. Figure 4 summarizes the results
attained by the different versions of ADMGA by
showing the configurations with and
values that
maximize the performance of each replacement
strategy.
The graphics show that RS 2 is capable of
outperforming standard ADMGA (RS 1) in the high
frequency scenarios. Replacement strategy 3, which
introduces mutated copies of the best individual in
the population, works well in the onemax problem,
but it is outperformed by the other strategies in most
of the dynamic scenarios based on the other two
functions. (RS 2 is 2-elitist, because this improves
its performance. Please note that RS 2 is quite
disruptive. This the payoff for having diversity
maintenance mechanisms, but the elitism guarantees
that the best solutions are not lost.)
Table 1: Kolmogorov-Smirnov tests (RS 2 vs RS1).
Results are shown as + signs when ADMGA with RS 2 is
significantly better than the ADMGA with RS 1, − when
RS 2 is significantly worst, and ≈ when the differences are
not statistically significant. Parameters as in fig. 4.
ε→ 600 1200 2400 4800 9600 19200 38400
onemax
+ + + +
≈ ≈ ≈
trap
+ + + + + + +
knapsack
+ + + + + + +
Table 1 summarizes the statistical tests conducted
on these results. RS 2 is compared with RS 1 using
Kolmogorov-Smirnov tests with 0.05 level of
significance. The tests show that RS 2 clearly
outperforms standard ADMGA (RS 1) in most of the
problems. The first objective of this study has been
accomplished: one of the schemes is able to improve
ADMGA’s performance in fast dynamic problems.
Let us now compare RS 2 with the other GAs.
Figure 5 compares ADMGA (RS 2) with GGA and
EIGA. As already stated, GGA and EIGA were
thoroughly tested in order to avoid unfair
comparisons. GGA works better with
1.0 and
2-elitism. Best population size is 16 for
onemax, and 30 for order-3 trap and knapsack
(same values were found for the remaining
algorithms). In general,GGA’s performance is
optimized by
1
⁄
except with knapsack, where
the best is
2
⁄
.
Table 2: Kolmogorov-Smirnov tests (RS 2 vs GGA). The
results are shown as + signs when ADMGA with RS 2 is
significantly better than GGA, − when RS 2 is
significantly worst, and ≈ when the differences are not
statistically significant. Parameters as in figure 5.
ε→
600
1200 2400 4800 9600 19200
38400
onemax
− − ≈ ≈ ≈ ≈ ≈
trap
≈ ≈
+ + + + +
k
napsack
− − ≈ ≈ ≈ + +
Figure 5 and table 2 shows that for 2400,
ADMGA is never outperformed by GGA. In
particular, the value above which ADMGA is at
least equivalent to GGA decreases from 4800 to 600
in order-3 trap (compare figures 3 and 5). Table 3
compares ADMGA with the standard strategy (RS
80
85
90
95
100
averagedofflineperformance
onemax
RS1
RS2
RS3
22
24
26
28
30
averagedofflineperformance
order‐3trap
RS1
RS2
RS3
1770
1790
1810
1830
averagedofflineperformance
knapsack
RS1
RS2
RS3
ICEC 2010 - International Conference on Evolutionary Computation
110
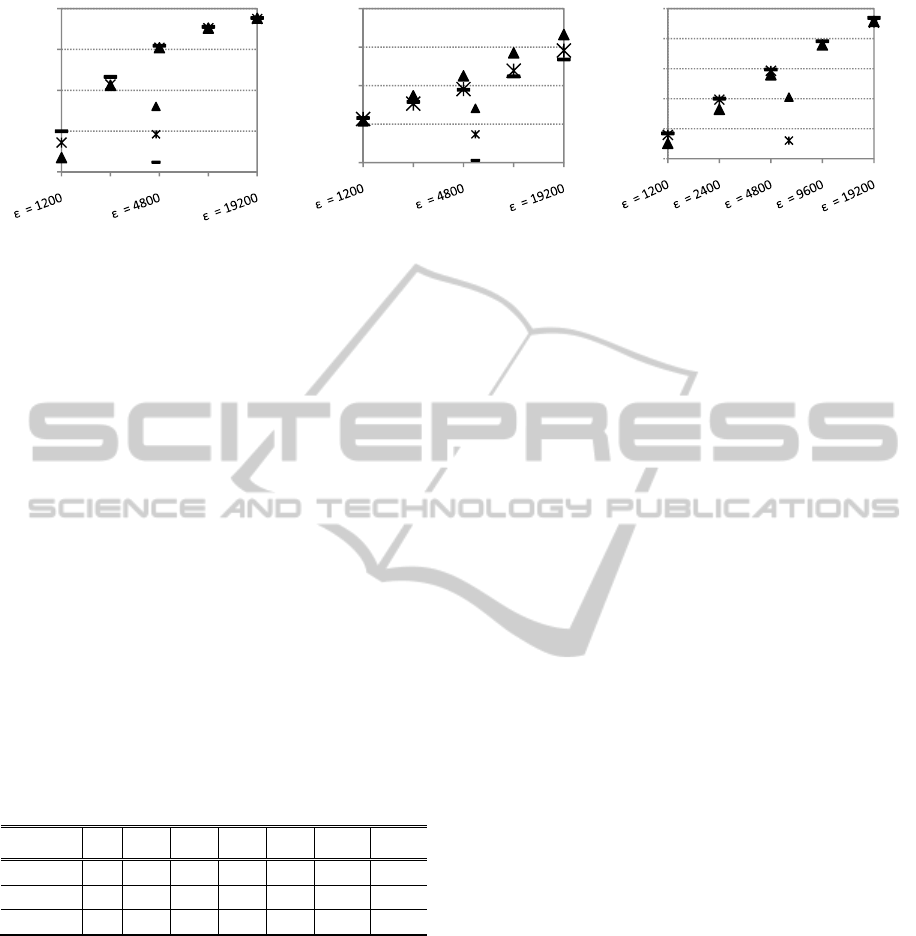
Figure 5: ADMGA (RS 2), GGA and EIGA. Parameters as in fig. 3 and 4. Population size n = 16 (onemax) and n = 30
(order-3 and knapsack). GGA with
1
⁄
(onemax and trap) and
2
⁄
(knapsack). EIGA with
2
⁄
and
0.2.
1) and GGA. By comparing tables 2 and 3, it is
noticeable that RS 2 reduces the above which
ADMGA is significantly better or at least
statistically equivalent to GGA in the dynamic
scenarios of the three base functions.
If we compare ADMGA’s replacement strategy 2
with EIGA the conclusions are similar: see figure 5
and table 4. EIGA performs better than ADMGA in
fast onemax problem and knapsack problems. On
the other hand, EIGA is outperformed by ADMGA
in almost every order-3 trap dynamic problem.
(Please note that in (Yang, 2008), EIGA is tested
with 1200 and 6000, a range that is
covered by the experiments conducted for this
paper).
Table 3: Kolmogorov-Smirnov tests (RS 1 vs GGA). The
results are shown as + signs when ADMGA with RS 1 is
significantly better than GGA, − when RS 1 is
significantly worst, and ≈ when the differences are not
statistically significant. Parameters as in figures 4 and 5.
ε→ 600 1200 2400 4800 9600 19200 38400
onemax
− − − − ≈ ≈ ≈
trap
− − − ≈
+ + +
knapsack
− − − − − ≈ +
As stated above, the comparisons in this study
were made considering the worst-case scenario, i.e.,
changes are hard to detect and a reliable detection
requires the reavaluation of the chromosomes that
are copied from previous generations. However, we
may consider a different assumption: changes are
easy to detect and all that is required is to evaluate
every old chromosomes after a change is detected.
Under these conditions, the results are different. The
summarized outcome of EIGA and ADMGA is
shown in Table 5: ADMGA clearly outperforms
EIGA in almost every dynamic problem. However,
at this point we cannot exclude the possibility of
population-wide elitism may now be biasing the
results towards ADMGA; therefore, other
experiments must be devised in order to properly
compare the GAs.
Figure 6 compares the genetic diversity, as
defined in (Fernandes, 2009), of the different
strategies. RS 2 is able to maintain diversity at a
higher level during the different periods. On the
other hand, the highly elitist strategy 3, as expected,
decreases the diversity when compared to the
standard strategy. These results may explain the
general behavior of the replacement strategies. Since
RS 2 is able to reduce diversity loss, it attains better
results throughout the test set.
6 CONCLUSIONS
This paper proposes new replacement schemes for
Adaptive Dissortative Mating Genetic Algorithm
(ADMGA). The main objective is to improve
standard ADMGA’s performance in dynamic
problems with high frequency of changes. One of
the proposed strategies outperforms the standard
strategy in most of the dynamic scenarios designed
to test the algorithms. This new strategy (RS 2)
simply mutates the chromosomes that remain in the
population after the recombination stage — the best
’ solutions in the parents’ population, where
is the population size and ’ is the offspring
population size — before reevaluating them.
The results show that ADMGA is capable of
outperforming not only a standard GA, but also the
Elitism-based Immigrants GA (EIGA) in some
classes of problems and dynamics: 1) when the
frequency of changes is lower, ADMGA is never
outperformed by the other GAs; 2) as for higher
frequencies, ADMGA is never outperformed by
80
85
90
95
100
averagedofflineperformance
onemax
ADMGA(RS2)
GGA
EIGA
22
24
26
28
30
order‐3trap
ADMGA(RS2)
GGA
EIGA
1780
1790
1800
1810
1820
1830
knapsack
ADMGA(RS
2)
GGA
INVESTIGATING REPLACEMENT STRATEGIES FOR THE ADAPTIVE DISSORTATIVE MATING GENETIC
ALGORITHM
111

GGA and EIGA in order-3 trap functions. Finally,
preliminary tests with non-stationary environments
in which the changes are easy to detect show that
ADMGA is able to outperform EIGA in every
(except one) scenario.
Table 4: Kolmogorov-Smirnov tests (RS 2 vs EIGA). The
results of the test are shown as + signs when ADMGA
with RS 2 is significantly better than EIGA, − when RS 2
is significantly worst, and ≈ when the differences are not
statistically significant. Parameters as in figure 5.
ε→
600
1200 2400 4800 9600 19200
38400
onemax
− − − ≈ ≈ ≈ ≈
trap
≈ ≈
+ + + + +
knapsack
− − ≈ ≈ ≈ ≈ ≈
One of ADMGA’s advantages over other GAs is
that it only requires two parameters that need to be
tuned ( and
), while EIGA, for instance, requires
the setting of four parameters (,
,
and ).
Since EIGA has been recently proposed as a GA
specifically conceived for dynamic optimization,
and since the report in (Yang, 2008) claims that the
algorithm performs well on dynamic, we may state
that ADMGA is a viable strategy for tackling
dynamic optimization problems.
Figure 6: RS 1, 2 and 3 genetic diversity. Dynamic order-3
trap function with 2400. Parameters as in figure 4.
Table 5: Kolmogorov-Smirnov tests (RS 2 vs EIGA). The
results of the test are shown as + signs when ADMGA
with RS 2 is significantly better than EIGA, − when RS 2
is significantly worst, and ≈ when the differences are not
statistically significant. Parameters as in figure 5.
ε→
600
1200 2400 4800 9600 19200
38400
onemax
+ + + + + +
≈
trap
+ + + + + + +
knapsack
+ + + + + + +
ACKNOWLEDGEMENTS
The first author wishes to thank FCT, Ministério da
Ciência e Tecnologia, his Research Fellowship
SFRH / BPD / 66876 / 2009, also supported by FCT
(ISR/IST plurianual funding) through the
POS_Conhecimento Program. This paper has also
been funded in part by the Spanish MICYT projects
NoHNES (TIN2007-68083) and TIN2008-06491-
C04-01 and the Junta de Andalucía P06-TIC-02025
and P07-TIC-03044.
REFERENCES
Angeline, P., 1997. Tracking Extrema in Dynamic
Environments. In Proc. of the 6
th
International Conf.
on Evolutionary Programming, Springer, 335-345.
Branke, J., 1999. Memory enhanced evolutionary
algorithms for changing optimization problems. In
Proc. of the 1999 IEEE Congress on Evolutionary
Computation, IEEE Press, 1875-1882.
Branke, J., Kaußler, T., Schmidt, C., Schmeck, H., 2000.
A multi-population approach to dynamic optimization
problems. In Proc. of the Adaptive Computing in
Design and Manufacturing (ACDM’2000), I.C.
Parmee, Ed., London, UK: Springer-Verlag, 299-308.
Branke, J., 2001. Evolutionary optimization in dynamic
environments. Norwel, MA: Kluwer.
Cobb, H. G., 1990. An investigation into the use of
hypermutation as an adaptive operator in GAs having
continuous, time-dependent nonstationary
environments. Technical Report AIC-90-001, Naval
Research Laboratory, Washington, USA.
Eschelman, L. J., Schaffer, J. D., 1991. Preventing
premature convergence in genetic algorithms by
preventing incest. In Proc. of the 4
th
International
Conference on Genetic Algorithms, Morgan
Kauffman, San Francisco, 115-122.
Fernandes, C. M., Rosa, A. C., 2001. A Study on Non-
Random Mating in Evolutionary Algorithms Using a
Royal Road Function. In Proc. of the 2001 Congress
on Evolutionary Computation, IEEE Press, 60-66.
Fernandes, C. M., Rosa, A. C., 2008. Self-adjusting the
intensity of dissortative mating of genetic algorithms.
Journal of Soft Computing, vol. 12, 955-979.
Fernandes, C. M., Rosa, A. C., 2008. Evolutionary
Algorithms with Dissortative Mating on Static and
Dynamic Environments. Advances in Evolutionary
Algorithms, W. Kosinski Ed., In-Tech, 181-206.
Fernandes, C. M., Merelo, J. J., Ramos, V., Rosa, A. C.,
2008. A Self-Organized Criticality Mutation Operator
for Dynamic Optimization Problems. In Proc. of the
2008 Genetic and Evolutionary Computation
Conference, ACM Press, pp. 937-944.
Fernandes, C. M., 2009. Diversity-enhanced Genetic
Algorithms for dynamic optimization. Ph.D Thesis,
0
0,1
0,2
0,3
0,4
0,5
0,6
0,7
0 4800 9600 14400 19200 24000
geneticdiversity
functionevaluations
order‐3trap
RS1
RS2
RS3
ICEC 2010 - International Conference on Evolutionary Computation
112

Tec. Univ. of Lisbon, (http://geneura.ugr.es/pub/
tesis/PhD-CFernandes.pdf)
Goldberg, D. E., Smith, R. E., 1987. Nonstationary
function optimization using genetic algorithms with
dominance and diploidy. In Proc. of the 2nd
International Conference on Genetic Algorithms, New
Jersey, 1987, 59-68.
Grefenstette, J. J., 1992. Genetic algorithms for changing
environments. In Parallel Problem Solving from
Nature II, R. Manner and B. Manderick, Eds., North-
Holland, Amsterdam, 137-144.
Ochoa, G., Madler-Kron, C., Rodriguez, R., Jaffe, K.,
2005. Assortative mating in genetic algorithms for
dynamic problems. In Proc. of the 2005
EvoWorkshops, LNCS 3449, Springer, pp. 617-622.
Ochoa, G., 2006, Error Thresholds in Genetic Algorithms.
Evolutionary Computation, 14(2), 157-182.
Ramsey, C. L., Grefenstette, J. J., 1993. Case-based
initialization of genetic algorithms. In Proc. of the 5
th
International Conference Genetic Algorithms, Morgan
Kaufmann, 84–91.
P. J. Russel, Genetics. Benjamin/Cummings, 1998.
Thierens, D., 1999. Scalability problems of simple GAs.
Evolutionary Computation 7(4), 331-352.
Tinós, R., Yang, S., 2007. A self-organizing random
immigrants GA for dynamic optimization problems.
Genetic Programming and Evolvable Machines 8(3),
255-286.
Yang, S., 2003. Non-stationary problem optimization
using the primal-dual genetic algorithm. In Proc. of
the 2003 IEEE Congress on Evolutionary
Computation, Vol. 3, 2246-2253.
Yang, S. Yao, X., 2005. Experimental study on PBIL
algorithms for dynamic optimization problems. Soft
Computing 9(11),815-834.
Yang, S., 2008. Genetic Algorithms with Memory- and
Elitism-Based Immigrants in Dynamic Environments.
Evolutionary Computation 16(3), 385-416.
INVESTIGATING REPLACEMENT STRATEGIES FOR THE ADAPTIVE DISSORTATIVE MATING GENETIC
ALGORITHM
113
