
LEAD DISCOVERY IN THE WEB
Iaakov Exman
1
and Michal Pinto
2
1
Software Engineering Dept.,
2
Pharmaceutical Engineering Dept.
Jerusalem College of Engineering, POB 3566, Jerusalem, 91035, Israel
Keywords: Lead, discovery, Lead-&-Search protocol, Knowledge representation, Linearized components.
Abstract: The Web is a huge and very promising source of medical drug leads. But, conventional search with generic
search engines, does not really obtain novel discoveries. Inspired by the drug discovery approach, we add
the idea of a "lead" to the search process. The resulting Lead-&-Search protocol avoids the trap of repeated
fruitless search and is domain independent. The approach is applied in practice to drug leads in the Web. To
serve as input to generic search engines, multi-dimensional chemical structures are linearized into strings,
which are sliced into keyword components. Search results are reordered by novelty criteria relevant to drug
discovery. Case studies demonstrate the approach: linearized components produce specific search outcomes,
with low risk of semantic ambiguity, facilitating reordering and filtering of results.
1 INTRODUCTION
Development of new medical drugs, from initial
concepts to commercial marketing, is an extremely
long process. Any path to shorten it is of great value.
Often, new drug development starts from leads –
molecules with desirable activity – later improved
by modifying groups of atoms (called fragments), to
increase activity and decrease undesirable side-
effects. We use the term component for fragment.
The Web is a huge potential source of leads to
new drugs. But, conventional search with a generic
engine, rarely results in unexpected discoveries.
Inspired by medical drug development, we added
the "lead" idea to the search process. One obtains a
generic Lead-&-Search protocol, applicable to any
Web discovery process, not just drugs.
The Lead-&-Search protocol alternates between
lead proposal and search phases, as needed, thereby
avoiding sterile repetitive search.
Our Web lead discovery approach is applied to
novel drug development. Much of the information
on potential drugs is multi-dimensional. This is an
obstacle to generic search engines limited to linear
keyword strings. We directly use linearized
structures sliced into components as search inputs.
2 LEAD-&-SEARCH – THE
PROTOCOL
Lead-&-Search is a completely generic protocol.
Though inspired by new drug development from
leads, it is applicable to any kind of Web discovery.
The Lead-&-Search protocol goal is to overcome
the problem of repeated fruitless search – analogous
to a local minimum in an optimization problem – by
random jumping to another lead. It aims to converge
at successful discovery results, by alternate steps of
proposing a lead and performing search.
The iterative Lead-&-Search process is
schematically depicted in Figure 2.1. Two spaces are
involved: a semantic space containing potential
leads – concepts used as inputs in a search engine
working in a search space – and the search space
containing result sets found in the Web.
The Lead-&-Search protocol starts from an
initial lead (Lead-1 in Fig. 2.1), as input to search.
The protocol should be independent of the initial
lead quality.
If search is fruitful, one may refine this Lead
with the acquired knowledge. If search does not
produce results as desired, one jumps to another lead
(Lead-2) and retries search.
The Lead-&-Search loop is repeated until it is
either successful or ends by a termination criterion.
The Lead-&-Search protocol is detailed in Fig. 2.2
in pseudo code. In order to apply the
471
Exman I. and Pinto M..
LEAD DISCOVERY IN THE WEB.
DOI: 10.5220/0003098304710474
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2010), pages 471-474
ISBN: 978-989-8425-28-7
Copyright
c
2010 SCITEPRESS (Science and Technology Publications, Lda.)
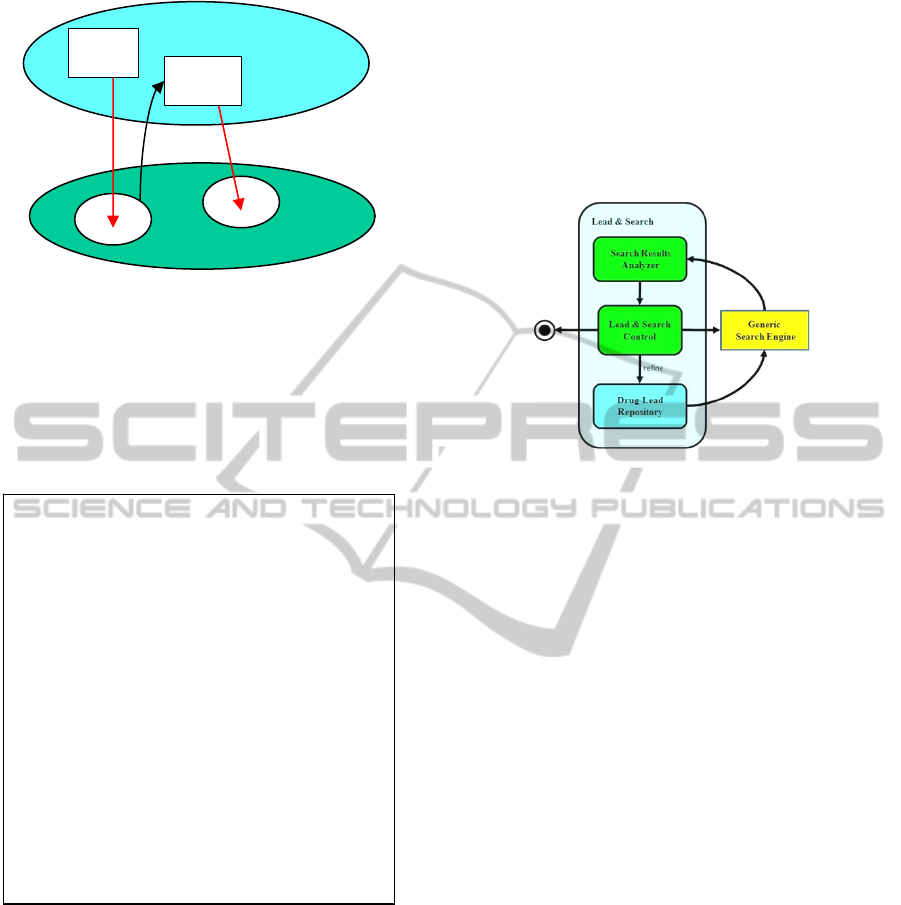
Figure 2.1: Schematic Lead-&-Search Protocol– It
alternates between proposing leads in a semantic space
and performing search in the search space, until it
succeeds or terminates.
Lead-&-Search protocol in practice one needs: a) to
set values to criteria (search-termination and lead-
timeout);
b)
to determine algorithms to choose and
refine a lead, perform search and random jumps.
Figure 2.2: Lead-&-Search Protocol in pseudo-code –
After an initiation, the loop alternates between search and
random jumps to other leads, if necessary.
3 LEAD & SEARCH SOFTWARE
ARCHITECTURE
The overall Lead & Search software architecture and
behavior is schematically seen in Fig. 3.1. A control
unit makes decisions, depending on the software
state and on current results. It decides whether to:
• Jump to another lead and perform search;
• Refine the drug-lead and repeat search;
• Terminate the whole process.
The Lead-&-Search software system interacts with
unmodified generic search engines. Search results
are reordered and filtered by an analyzer module.
The drug-lead repository provides input to the
search engine. Lead-&-Search success means:
1. The drug-lead is gradually refined;
2. New leads are discovered.
Figure 3.1: Lead-&-Search Software Behavior Units – The
control module decides whether to jump to another lead
and search, to refine the lead, or to end the whole process.
4 SEARCH – WITH LINEARIZED
STRUCTURES
Generic search engines are powerful, but only accept
linear input strings. An important issue is which
kinds of information can be effectively used in this
way; see (Konyk, 2008) and (Searls, 2005).
Medical drugs are based on an active
substance, represented by a molecule. There is a
variety of knowledge types about a substance:
• Text – substance name and properties;
• Images – diverse physical spectra;
• 2-Dimensional (2-D) formula;
• 3-Dimensional (3-D) model.
Text does not deserve special considerations.
Spectral information images say, NMR (Nuclear
Magnetic Resonance) see e.g. (Homans, 2004), are
too complex to serve as direct search inputs.
Molecular structures – 2-D or 3-D – are graphs
with edges connecting discrete entities (atoms or
groups of atoms). They are amenable to linearized
forms useful as search inputs.
SMILES is a "Simplified Molecular Input Line
Entry" proposed by Weininger et al. (
Weininger
,
1988), (Weininger, 1989). It is an unambiguous
string describing 2-D or 3-D structures. There is an
open standard (OpenSMILES, 2007).
SMILES is obtained by depth-first tree traversal
of the molecule graph. The graph is trimmed
L
ead-&-
S
earch – Protocol
//Initiation phase
Get initial-Lead; Set Current-Lead = initial-
Lead;
//Loop
While (Search-Termination = False)
Do {While (Lead-Timeout = False)
Do {Perform Search;
If Search-Termination-Criterion Achieved
Then Set Search-Termination = True;
Return;
If Lead-Timeout-Criterion Reached
Then Set Lead-Timeout = True;
Else Refine Current-Lead;}
If Lead-Timeout = True
Then {Random Jump to Another-Lead;
Set Current-Lead = Another-Lead;
Set Lead-Timeout = False
;}
}
Semantic Space
Search
S
p
ace
Lead-1
Search
Region-1
Search
Region-2
Lead-2
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
472

(removing hydrogen atoms implicit by the graph
skeleton); cycles are broken obtaining a spanning
tree (numeric suffixes mark linked nodes of broken
cycles). Parentheses mark tree branching points.
5 VALIDATION
Preliminary validation is provided by case studies
with different types of physiological activity. Search
with sliced linearized components obtains drugs and
potential leads among the results.
5.1 Case Study 1: Vancomycin
Vancomycin is an antibiotic of molecular formula
C
66
H
77
C
l2
N
9
O
24
. Fig. 5.1 displays its SMILES string
(the bold [red] component is the search input). The
corresponding 2-D structure is seen in Fig. 5.2.
Table 1 shows search results for three search
engines (bing, google and yahoo). For the small
result sets obtained (between 15 and 30 results) there
are relatively many potential substances of interest.
Figure 5.1: Vancomycin SMILES Linear String –The
fragment in bold [red] was used to search potential leads.
Table 1: Vancomycin Search Results.
# Drug/Lead
Search
rank
notes
1 Chloro-
orienticin
bing/2 Glycopeptide Antibiotic
2 telavancin google/
3
Semisynthetic
Vancomycin
3 methicillin google/
9
Not used anymore
4 chloro-
eremomyci
n
yahoo/
19
Vancomycin family
antibiotic (synonym to 1)
Result Set sizes: bing = 15; google = 28; yahoo = 30; Search rank
format is: "engine name / numeric rank of result", say "bing/2".
5.2 Case Study 2: Midazolam
Midazolam, a hypnotic-sedative drug, has molecular
formula C
18
H
13
ClFN
3.
Fig. 5.3 displays a SMILES
string, corresponding to the 2-D structure in Fig. 5.4.
Figure 5.2: Vancomycin 2-D Structure – It shows atoms
(say O=Oxygen), groups of atoms (CH
3
=methyl), linked
by chemical bonds (graph edges), and hexagonal rings.
Figure 5.3: Midazolam SMILES String –The fragment in
bold [red] was used to search potential leads.
Figure 5.4: Midazolam 2-D Structure – It shows
hexagonal and other rings and atoms (N=Nitrogen,
F=Fluor, Cl=Chlorine), linked by chemical bonds.
The component C2N1C3=C(C=C(C=C3)Cl)
stressed in bold in Fig. 5.3 served as search input.
Results are seen in Table 2 for the same engines.
The result sets (between 11 and 138 results) are still
relatively small. The number of potential substances
of interest is of the same order of magnitude.
Table 2: Midazolam Search Results.
# Drug/Lead
Search
rank
notes
1
deracyn yahoo/1 generic name: adinazolam
2
4-hydroxy-
alprazolam
bing/1,
google/2
analog of alprazolam
3
alprazolam google/4
sedative to treat insomnia
Result Set sizes: bing = 11; google = 138; yahoo = 41;
5.3 Case Study 3: Nelarabine
Nelarabine is a chemoterapy drug used to treat
leukemia. Its molecular formula is C
11
H
15
N
5
O
5.
Fig.
CC1=NC=C2N1C3=C(C=C(C=C3)Cl)C(=NC2)C4=CC=
CC=C4F
CC1C(C(CC(O1)OC2C(C(C(OC2OC3=C4C=C5C=C3OC6
=C(C=C(C=C6)C(C(C(=O)NC(C(=O)NC5C(=O)NC7C8=
CC(=C(C=C8)O)C9=C(C=C(C=C9C(NC(=O)C(C(C1=CC(
=C(O4)C=C1)Cl)O)NC7=O)C(=O)O)O)O)CC(=O)N)NC(=
O)C(CC(C)C)NC)O)Cl)CO)O)O)(C)N)O.Cl
LEAD DISCOVERY IN THE WEB
473
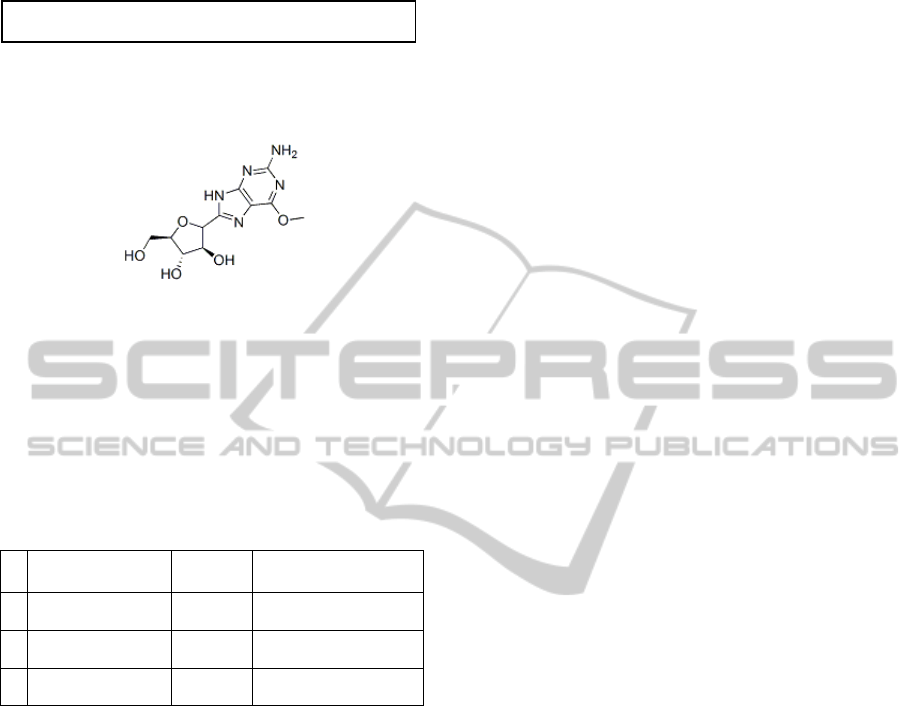
5.5 displays a SMILES string for this substance. Fig.
5.6 shows its 2-D structure.
Figure 5.5: Midazolam SMILES String – This is a linear
string representing Nelarabine. The fragment in bold [red]
was used to search potential leads.
Figure 5.6: Nelarabine 2-D Structure – This formula
displays hexagonal and pentagonal rings and atoms and
groups (say NH
2
=Amino), linked by chemical bonds.
The component COC1=NC(N)=NC2=C1N=C
highlighted in bold [red] in Fig. 5.5 served as search
input. Result sets (249 up to 429 results) seen in
Table 3 are manageable. The number of substances
of interest is of the same order of magnitude.
Table 3: Nelarabine Search Results.
# Drug/Lead
Search
rank
notes
1
6-O-Methyl
Guanosine
yahoo/1 guanine derivative
used in drug design
2
alfuzosin google/2 treats benign prostatic
hyperplasia
3
6-methoxy-9-
methyl-9H-purine
yahoo/7 substance for drug
design
Result Set sizes: bing = 429; google = 276; yahoo = 249;
6 DISCUSSION
Some preliminary conclusions from case studies are:
a) one can control the jump size between
consecutive leads in the Lead-&-Search
protocol, by controlling the leads' overlap;
b) randomly
sliced SMILES strings give small
result sets, being improbable combinations of
letters; the risk of semantic ambiguity is low;
c) one expects an approximately inverse
proportional relation between components'
string size and result set size;
d) direct search of drug names, say Vancomicyn,
is too weak to be of value for discovery.
The Lead notion has been used within a closed
computational framework (Wise, 1983) and (Exman,
1988), but has not been yet applied to the Web.
Another linear molecular naming system is
InChI (
McNaught 2006). SMILES is more readable
than InChI, but conveys less information. Our choice
of SMILES can be changed, if proved necessary.
In order to demonstrate the actual efficiency of
the approach for drug discovery, an extensive
investigation of a variety of drug families is needed.
6.1 Main Contribution
Our main contribution is the randomized "Lead"
proposal phase added to "Search", forming the
"Lead-&-Search" protocol, a powerful discovery
mechanism in the Web
REFERENCES
Exman, I. and D. H. Smith – "Get a Lead & Search: A
Strategy for Computer-Aided Drug Design'', in Symp.
Expert Systems Applications in Chemistry, ACS, 196
th
National Meeting, Los Angeles, p. COMP-69, (1988).
Homans, S. W., “NMR Spectroscopy Tools for Structure-
Aided Drug Design”, Angewandte Chemie Int. Ed.
Vol. 43, pp. 290–300, (2004).
Konyk, M., A. De Leon and M. Dumontier, "Chemical
Knowledge for the Semantic Web", in A. Bairoch, S.
Cohen-Boulakia, and C. Froidevaux (eds.): DILS,
LNBI 5109, pp. 169-176, Springer, Berlin (2008).
McNaught, Alan, "The IUPAC International Chemical
Identifier: InChI", Chemistry Int., Vol. 28 (6) (2006).
OpenSMILES Standard – http://www.opensmiles.org/
Draft (November 2007).
Searls, D. B., "Data integration: challenges for drug
discovery", Nature Reviews Drug Discovery 4, 45-58
(January 2005).
Weininger, D., "SMILES, a chemical language and
information system. 1. Introduction to methodology
and encoding rules", J. Chem. Inf. Comput. Sci. Vol.
28. pp. 31-36 (1988).
Weininger, D., Weininger, A., Weininger, J.L. "SMILES.
2. Algorithm for generation of unique SMILES
notation", J. Chem. Inf. Comput. Sci, 29, pp. 97-101
(1989).
Wise, M., R. D. Cramer, D. Smith and I. Exman -
“Progress in 3-D Drug Design: the use of Real Time
Colour Graphics and Computer Postulation of
Bioactive Molecules in DYLOMMS" – in J. Dearden,
(ed.) Quantitative Approaches to Drug Design, Proc.
4
th
European Symp. on "Chemical Structure-
Biological Activity: Quantitative Approaches". Bath
(U.K.), pp. 145-146., Elsevier, Amsterdam, 1983.
COC1=NC(N)=NC2=C1N=CN2C1OC(CO)C(O)C1O
KDIR 2010 - International Conference on Knowledge Discovery and Information Retrieval
474
