
ROBUSTNESS OF EXON CGH ARRAY DESIGNS
Tomasz Gambin
1
, Pawel Stankiewicz
2
, Maciej Sykulski
3
and Anna Gambin
3,4
1
Institute of Computer Science, Warsaw University of Technology, 15/19 Nowowiejska, 00-665 Warsaw, Poland
2
Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, U.S.A.
3
Institute of Informatics, University of Warsaw, 2 Banacha, 02-097 Warsaw, Poland
4
Mossakowski Medical Research Centre Polish Academy of Sciences, 5 Pawinskiego, 02-106 Warsaw, Poland
Keywords:
aCGH, Segmentation, Noise robustness, Design optimization, DNA copy.
Abstract:
Array-comparative genomic hybridization (aCGH) technology enables rapid, high-resolution analysis of ge-
nomic rearrangements. With the use of it, genome copy number changes and rearrangement breakpoints can
be detected and analyzed at resolutions down to a few kilobases. An exon array CGH approach proposed
recently accurately measures copy-number changes of individual exons in the human genome. The crucial
and highly non-trivial starting task is the design of an array, i.e. the choice of appropriate (multi)set of oligos.
The success of the whole high-level analysis depends on the quality of the design. Also, the comparison of
several alternative designs of array CGH constitutes an important step in development of new diagnostic chip.
In this paper we deal with these two often neglected issues.
We propose new approach to measure the quality of array CGH designs. Our measures reflect the robustness of
rearrangements detection to the noise (mostly experimental measurement error). The method is parametrized
by the segmentation algorithm used to identify aberrations. We implemented the efficient Monte Carlo method
for testing noise robustness within DNAcopy procedure. Developed framework has been applied to evaluation
of functional quality of several optimized array designs.
1 INTRODUCTION
DNA copy number aberrations that cause a gain or
loss of chromosomal material are associated with
many types of genomic disorders like mental retar-
dation, congenital malformations or autism (Lupski,
2009; Shaw et al., 2004). Moreover, genetic aber-
rations are characteristic of many cancer types and
are thought to drive some cancer pathogenesis pro-
cess (O’Hagan et al., 2003; Snijders et al., 2005;
Wang et al., 2006; Lai et al., 2007).
Array comparative genomic hybridization
(aCGH) became the standard protocol for identifying
segmental copy number alterations in disease state
genomes (Pollack et al., 1999; Perry et al., 2008).
In typical experiment each DNA (e.g. diseased
patient vs. healthy donor, or normal tissue vs. tumor)
is labeled by different fluorescent dye, and then
hybridized to an array. Signal fluorescent intensities
of each spot from both samples are considered to be
proportional to the amount of respective genomic
sequence present.
One can classify the CGH arrays into two types.
The first kind, targeted CGH arrays provide high-
resolution coverage of the genome primarily in areas
containing known, clinically significant aberrations,
see e. g. (Thomas et al., 2005; Caserta et al., 2008).
The second kind, whole-genome arrays, provide high
resolution coverage of the entire genome (Barrett
et al., 2004). However in many applications the
design of the array should combine these two ap-
proaches: the exploration of the whole genome with
the special focus on some specific regions (e.g. con-
taining genes related to the disease under study).
Related Research. The array design is the starting
point of the study on genomic disorders underlying
a given disease (Lemoine et al., 2009). There is a
large body of research concerning array design task,
see e.g. (Lipson et al., 2002; Lipson et al., 2007).
Similarly many papers consider the issues of normal-
ization and detrending array CGH data (Chen et al.,
2008; van Hijum et al., 2008; Staaf et al., 2007; Kreil
and Russell, 2005).
However, while conducting the large-scale
173
Gambin T., Stankiewicz P., Sykulski M. and Gambin A..
ROBUSTNESS OF EXON CGH ARRAY DESIGNS.
DOI: 10.5220/0003153201730182
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2011), pages 173-182
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

biomedical research projects it is a reasonable
practice to provide several prototype array designs.
A matter of fundamental importance here is how to
compare the functional quality of different arrays
to choose the best one for further experiments.
Moreover, often for this comparison task researchers
dispose of only limited amount of experimental data.
In contrast to array design and normalization stud-
ies, there are only few approaches proposed so far
in the literature to the problem of comparison be-
tween different array designs. There are some stan-
dard statistics calculated for purpose of array com-
parison. They comprise usually: Signal to Noise Ra-
tio, Derivative Log Ratio Standard Deviation, Back-
ground Noise, etc (Carter, 2002). In (Coe et al., 2007)
to compare the resolution of different arrays the new
performance measure called ”functional resolution”
was proposed. This measure incorporates the unifor-
mity of element spacing on the array and the sensitiv-
ity of the array to single-copy alterations.
Our Results. Analogously to other high-
throughput technologies (like mass spectrometry
or expression microarrays) various sources of tech-
nical and biological variation affect the array CGH
experiment. The measurement noise comes from
the preparation of the microarray slide and the
hybridization process, while the biological variability
arises from the heterogeneity of the cells in the
inspected samples (e.g. mosaicism (Iourov et al.,
2008)). However, despite increasing resolution of
CGH arrays the variation in signal measurements
cannot be eliminated. Therefore the methods capable
to detect aberrations even in very noisy data are of
great interest. Most of proposed solutions rely on
so-called segmentation methods that try to divide the
data into segments representing aberrant and normal
regions (Cahan et al., 2008; Daz-Uriarte and Rueda,
2007; Ben-Yaacov and Eldar, 2008; Lipson et al.,
2006).
According to several comparative studies pub-
lished so far (Willenbrock and Fridlyand, 2005) one
of the best performing method for finding copy num-
ber segments is Circular Binary Segmentation (CBS),
a segmentation approach based on finding change-
points in data (implemented e.g. in DNAcopy (Olshen
et al., 2004) R package).
Our goal in this study was to develop the frame-
work for performance comparison of different CGH
array designs. We decided to explore the concept of
robustness. The proposed methodology follows the
general concept of robust statistics (Hampel et al.,
2005), quoting B.D. Ripley an important area that is
used a lot less than it ought to be.
In our approach we consider the design robust
when it is effective in the detection of aberrations in
the presence of noise. The segmentation obtained for
the given design is treated here as a robust estimator
of rearrangement regions. Better designs correspond
to more robust estimators, i.e., those approximating
the aberrations for the data contaminated with the
noise. To our best knowledge, this work is the first
method that uses the noise sensitivity of segmentation
algorithm to compare different array designs. Aiming
in testing the robustness of a design we enhance the
DNAcopy method by incorporating parametrized
noise model. The R package named DNAcopyNoise
is provided as supplementary material available at
http://bioputer.mimuw.edu.pl/software/DNAcopyNoise.
Our results are twofold: firstly, using synthetic
data we demonstrate the usefulness of robustness
measure for array performance comparison. Sec-
ondly, we apply the concept of robustness to select
the best one from several optimized designs. The op-
timization aimed in reducing array size while keeping
the same rearrangements detection ability.
Organization of the Paper. Section Methods con-
tains the description of datasets used in our experi-
mental study. We decided to test our method on syn-
thetic datasets representing designs of different qual-
ity. Then we present the 180 K exon array design.
The enhancement of DNAcopy package is presented
and our performance quality measures are defined. In
the Results Section we present the evaluation of our
measure for hybridization experiments and robustness
based comparison of optimized designs. In Conclu-
sions we summarize our approach and sketch further
developments.
2 METHODS
2.1 Synthetic Array Design
Aiming in validation of robustness approach we gen-
erate several datasets using framework from (Willen-
brock and Fridlyand, 2005). Two types of datasets
generators are considered: they correspond to differ-
ent genomic rearrangements structure (high density of
relatively short segments, like in cancer tissues versus
rare long aberrant segments characteristic to genomic
disorders). For each type of data we consider differ-
ent array designs. E.g., for data of first type, dataset
(a) presented in Figure 1 is the exemplary output of
aCGH experiments performed on well designed array.
Dataset (b) corresponds to experimental data from the
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
174
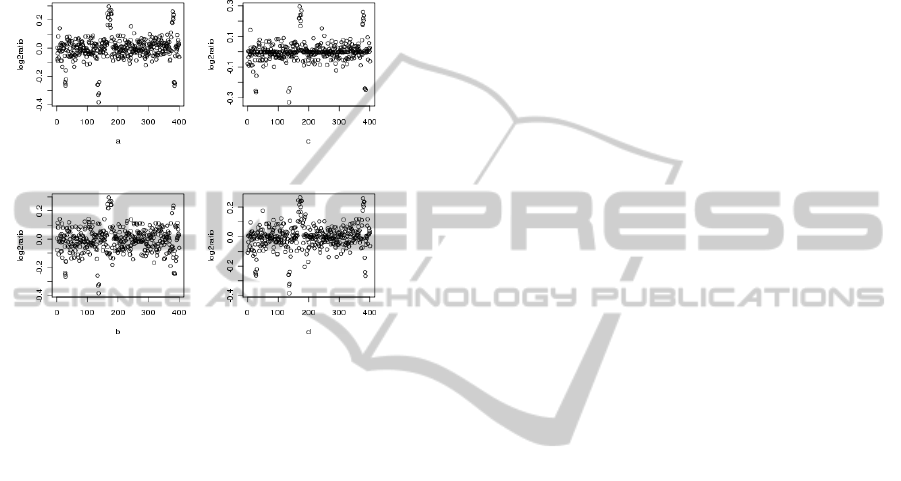
design, in which the inappropriate probe selection re-
sulted in poor hybridization. The generator (b) is ob-
tained as the following modification of the original
generator (a). We choose uniformly at random 20
percent of probes and multiply their signal intensity
by the coefficient sampled from beta distribution with
shape parameters α = 2 and β = 20 (unimodal distri-
bution defined on the interval [0, 1]).
Figure 1: Plots show log2ratio (y-axis) vs. genomic location
(x-axis) for synthetic datasets corresponding to four differ-
ent array designs: (a) original datasets, (b) dataset with sim-
ulated poor hybridization effect, (c) dataset with simulated
error-prone analysis procedures, (d) dataset with both ef-
fects.
The generator corresponding to array design (c)
mimics the problems arising from erroneous anal-
ysis protocol that results in significant background
noise. We assume here, that some probes may be er-
roneously analysed already during the scanning pro-
cess and only one from Red (cy5) and Green (cy3)
signal is detected. To model such situation we choose
uniformly at random 15% of probes and sample their
intensities from the beta distribution with parameters
α = 0.7 and β = 0.7. Such readouts correspond to
the probe signals not well scattered around zero in the
typical MA plot. The design (d) suffers from both
shortcomings. We generate 40 datasets using each de-
sign. One synthetic genome hybridization experiment
measure the signal intensities of 10000 probes located
on 10 chromosomes.
2.2 Exon CGH Array Design
Our new design quality measure has been tested on
samples obtained in aCGH experiments. The dataset
come from 60 arrays hybridized with DNA from sub-
jects with epilepsy, autism, heart defects and men-
tal disorders. Each experiment was performed on the
180 K exon targeted oligonucleotide array.
Prototype Design. The design of the chip involved
two stages. First, the prototype covering only ex-
onic and microRNA regions was constructed. The
main aim at this stage was to develop the array that
allows detecting DNA copy number changes of the
single exon. Therefore, it was postulated to cover
each exon by the same number of oligos. For a given
set of 1714 selected genes (including those related to
epilepsy, autism, heart defects, mental disorders and
other known pathologies) it was decided that each
exon would be covered by approximately 6 probes.
Cleaning Stage. The prototype coverage was two
times denser than the desired one in the final ver-
sion. A set of hybridizations was performed with the
prototype version. Performance score of each probe
was computed as following: segmentation was per-
formed on data from these experiments. Let us call
the empirical cumulative distribution function for dis-
tribution of logratios deviations from their segments
means F . The distribution F was estimated from
all experiments from the prototype version. For each
probe we perform two sided Kolomogorov-Smirnov
(K-S) test comparing the logratio deviation from seg-
ment mean with distribution F . We assign the p-value
obtained in this test as a score of the probe.
Next step involved combining the prototype de-
sign with backbone, i.e., probes putted uniformly
across the genome. Densely covered regions, ex-
onic double covered regions were thinned with heuris-
tic approach which considered previously assigned
scores and uniformity of nascent coverage (sizes of
introduced gaps).
2.3 Enhancement of DNACopy
DNAcopy package for R environment implements
circular binary segmentation algorithm (Olshen et al.,
2004). CBS algorithm finds segmentation by recur-
sively splitting subsequent segments into three, or two
smaller ones. Each segment cut is found by maximiz-
ing the following statistic:
Z
C
= max
1≤i< j≤n
t
i j
(1)
where t
i j
is t-statistic for probes resulting from parti-
tion of the cyclic logratio series at points i, j into two
samples: probes inside the interval (i,j), and its com-
plement.
Segmentation proceeds when the null hypothesis
is rejected, that is when Z
C
is above upper α−quantile
of null distribution Z
∗
C
.
ROBUSTNESS OF EXON CGH ARRAY DESIGNS
175

CBS algorithm estimates the null distribution with
the use of permutation method and tail probability es-
timation.
To estimate robustness of a segment we introduce
a Gaussian noise to the logratio data. We are inter-
ested in finding minimal level of noise that is very
likely to make the considered segment undetectable,
i.e., the maximal level that still guarantees that seg-
ment persists. Detecting these numbers through sim-
ulation requires extensive sampling since the intro-
duced noise is highly dimensional random variable.
To avoid running CBS algorithm many times, we in-
troduced the noise inside the sampling phase. CBS
use sampling to estimate the null distribution, by per-
mutation method. In our algorithm, every permuta-
tion is sampled with random noise added with zero
mean and η standard deviation. This changes the Z
∗
C
distribution and the sought quantile. This is com-
pared with the previously computed, however scaled
accordingly to introduced noise variance, t
i j
statistic
for the analyzed segment.
By tuning CBS parameters, specifically by, in-
creasing the number of permutations in each step, the
answer we obtain (if the segment is detectable with in-
troduced noise level η) is statistically significant. To
assign η
k
to each aberrant segment k we follow the
original, not noisy, CBS segmentation sequence, and
introduce noise in binary search fashion up to desired
precision.
2.4 Robustness Measure
It is inevitable that the measurement precision vary
considerably between probes depending on the hy-
bridization efficiency. Hence some regions of the
genome are analyzed with significantly higher preci-
sion than others (Baldocchi et al., 2005). Therefore
it is desirable to model the effectiveness of specific
array region in detecting aberrations. We propose an
approach that allows to evaluate the quality measure
for a whole array but also to focus on specific set
of probes. In our method we measure the quality of
array design using noise robustness of segmentation
algorithm performed for all accessible aCGH experi-
ments.
The intuition behind this approach can be ex-
plained in simple terms. Segmentation algorithm pro-
vides the information about comparative hybridiza-
tion experiment. Aberrant segments are easily de-
tectable if they are represented by good quality
probes. Good probes should tolerate higher level of
measurement noise than poor quality probes. There-
fore we conduct segmentation procedure for several
increasing noise levels and observe the behavior of
aberrant segments. There is certain number of seg-
ments found for original experimental data. Then
we simulate some measurement noise and repeat seg-
mentation algorithm. Some segments (consisted of
poor quality probes) disappear and we continue this
process, memorizing for each segment the maximal
noise level, for which this segment is still identifi-
able (for a fixed segment k we denote this value by
η
k
). The output of several segmentation stages for 2
different (synthetic) designs is presented in Figure 2.
Clearly, the left panel corresponds to more robust de-
sign.
Let us fix the aCGH experiment and let η
k
denote
the noise level of the maximal noise resistance of kth
segment defined as above. The level of noise is mea-
sured with reference to baseline variation (standard
deviation of probes in non aberrant regions). The ro-
bustness of probe k is defined as:
θ
k
=
η
k
length(k) · |mean(k)|
(2)
where length(k) is the length of segment k (measured
in the number of probes), and |mean(k)| is the abso-
lute value of mean of signal intensities along the seg-
ment. We assign the segment robustness to all the
probes it contains.
Now we combine the segmentation robustness of
several aCGH experiments into the measure of array
design quality. The robustness score for an array is
composed from robustness of probes it consists of.
Note that, we can estimate the quality only for those
probes that are witnesses of some aberration. Con-
sider a single probe k and assume, that it belongs to
aberrant segment in some samples (according to seg-
mentation algorithm run for original data). To this
probe robustness scores θ
i
1
k
, θ
i
2
k
, . . . θ
i
m
k
have been as-
signed in experiments i
1
, . . . i
m
. Assume, that there are
m accessible experiments in total. As an overall qual-
ity of this probe we can take the median of the empir-
ical distribution of robustness scores θ
i
1
k
, θ
i
2
k
, . . . θ
i
m
k
.
However in the case of limited number of acces-
sible experimental data we encounter here the prob-
lem of insufficient statistic, because a single probe
can be the witness of only few aberrations. To avoid
this difficulty we apply the sliding window approach.
The empirical distribution of probe robustness is com-
posed for all probes contained in the window of pre-
defined length n (depending on the resolution of an
array). The median of this distribution is calculated
yielding the smoothed version of the overall probe
quality.
The next neighboring window is shifted by the
half of the window length. Therefore any single
probe contributes to exactly two window statistics
(the boundary probes are ignored). Assume that the
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
176
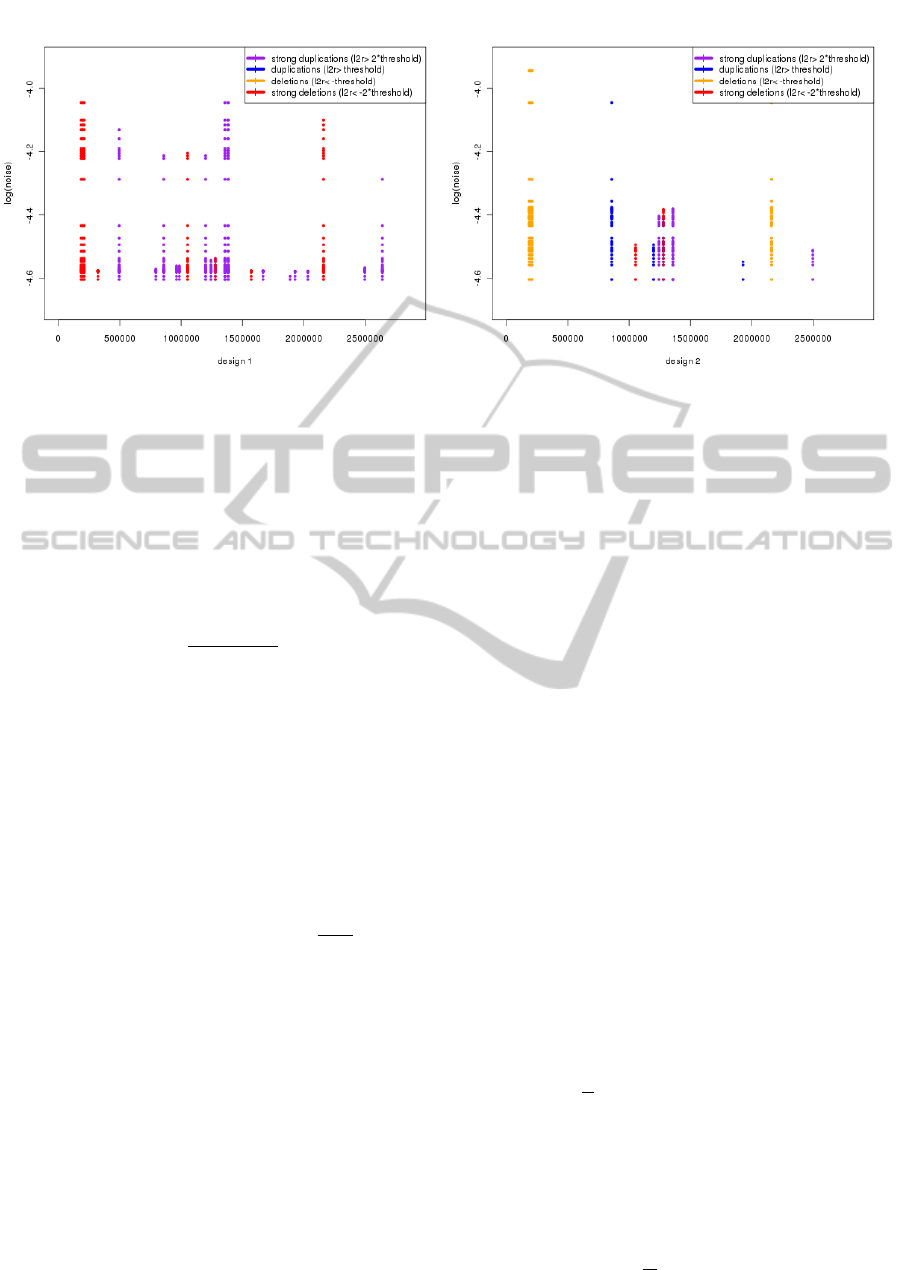
Figure 2: The resistance of aberrant segments for increasing noise. y-axis correspond to increasing noise level (logarithmized),
different segments are placed along x-axis (genomic location), the logratios are color-coded.
median (µ
L
) from the first window is calculated for i
L
events (aCGH experiments in which this probe lies in
the aberrant segment) and the second µ
R
for i
R
events.
Then the ith probe robustness for the array A is de-
fined as:
Θ
A
i
=
i
L
µ
L
+ i
R
µ
R
i
L
+ i
R
(3)
The robustness of array design A (containing N
probes) can be calculated by taking the average ro-
bustness of all probes.
However, the important issue here is that the cal-
culation of robustness for some probes relies on many
detected aberrations containing this probe, while for
others the robustness measure is supported by only
few witnesses. Consider once more the probe i and
two windows containing it. A support for the ith
probe robustness Θ
A
i
is defined as s
A
i
=
i
L
+i
R
nm
i.e., the
percent of experiments in which this probe or its sur-
rounding probes are witnesses of some aberration.
The support vector is composed of all probe sup-
ports s
s
s
A
= s
A
1
, . . . , s
A
i
, . . . , s
A
N
. This vector is further
transformed into importance weights vector ω
ω
ω
A
=
ω
A
1
, . . . , ω
A
N
by appropriate normalization and scaling
(the scaling function flatten out the support vector, as
higher support values have roughly the same impact).
Finally, the robustness of array design A is defined as:
Θ
A
= Σ
i
ω
A
i
Θ
A
i
(4)
In the next Section plots illustrating the robustness for
all probes use logarithmic scale for Θ
A
i
.
2.5 Optimizing Exon CGH Array
Design via Relative Robustness
The robustness measure Θ
A
defined for a given ar-
ray design A allows to estimate the functional per-
formance of A i.e., the efficiency of rearrangements
detection for noisy data. In this section we study the
problem of array design optimization. Our goal is to
eliminate certain percent of probes to obtain smaller
design which has comparable performance.
Here we assume that the segmentation Π found
for the original design reflects the real genomic aber-
rations. We refer to segmentation Π while measuring
the robustness of smaller designs. We compare the
optimized array with the original one looking at its
segmentation’s evolution for increasing noise level.
Let us fix the noise level η and define the distance
between two segmentations (say the original Π and
another one Π
i
) σ
η
(Π, Π
i
) similarly to raw distance
in (Liu et al., 2006), i.e., if both samples have a gain
(or loss) at the same genomic interval τ we consider
them identical, otherwise this genomic interval con-
tributes to the total distance. The contribution from
single interval is defined as its length (measured in
nucleotides) divided by the length of whole genome
(Γ), i.e.:
σ
η
(Π, Π
i
) =
1
Γ
∑
τ:τ differs between Π and Π
i
length(τ) (5)
To calculate the total distance σ
tot
η
we sum up the
contributions for all genomic intervals that differ be-
tween two samples and take the average over all m
experiments.
σ
tot
η
=
1
m
m
∑
i=1
σ
η
(Π, Π
i
) (6)
ROBUSTNESS OF EXON CGH ARRAY DESIGNS
177
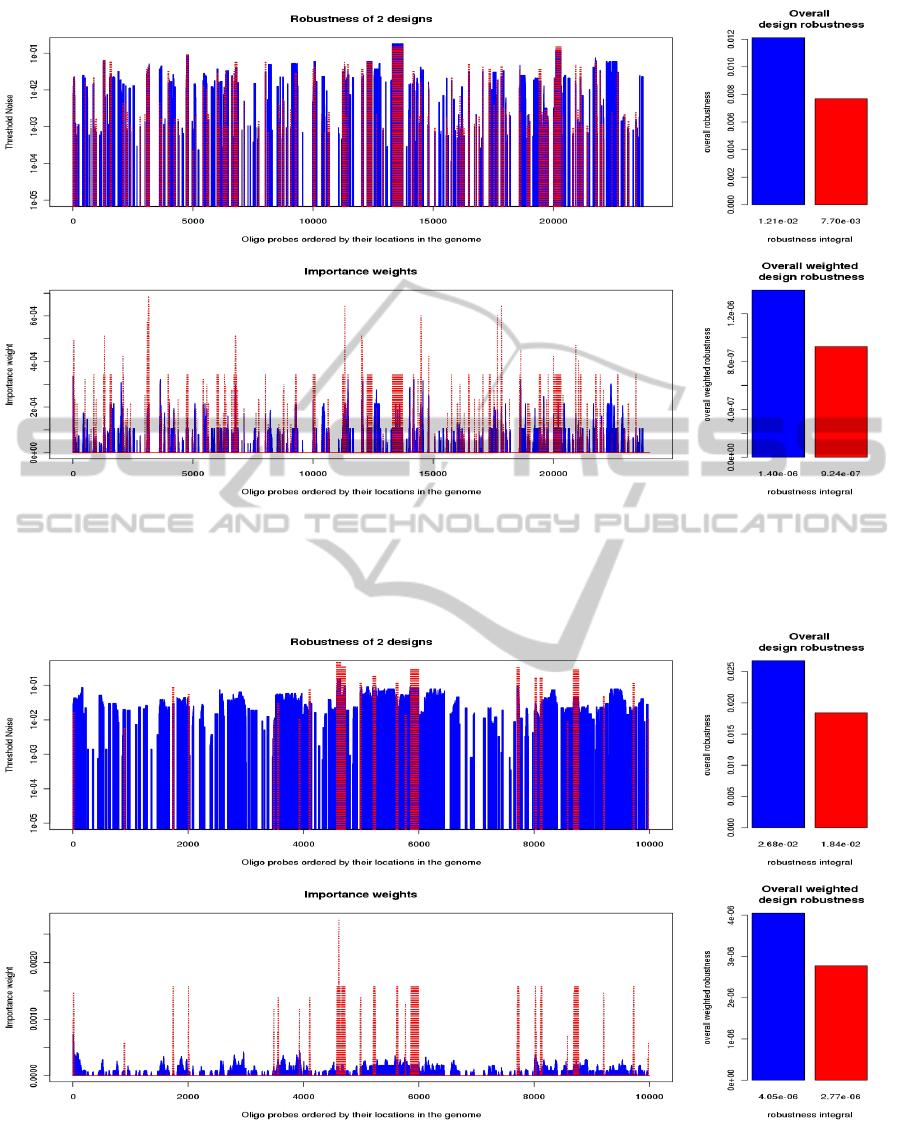
Figure 3: The robustness compared for two synthetic designs. The robustness has been calculated for all probes (upper plot)
as well as corresponding weights importance (lower plot). The structure of genomic rearrangements mimics the abnormalities
in cancer cells. Good design is coded in blue. Red design contains 20% of poorly hybridizing probes and 15% of outliers
(probes causing erroneous scanning).
Figure 4: The robustness compared for two synthetic designs. The robustness has been calculated for all probes (upper plot)
as well as corresponding weights importance (lower plot). The structure of genomic rearrangements mimics the abnormalities
in classical genetic disorder (relatively rare long aberrant segments). Good design is coded in blue. Red design contains 15%
of outliers (probes causing erroneous scanning).
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
178

Figure 5: Comparison of segmentations on optimized and original designs. Figure (a) shows the part of logratio data (1000
oligos on chr 15 - x-axis) obtained in 60 aCGH experiments (y-axis). There can be seen some common copy number changes
for all experiments (probably CNV’s), e.g., small duplications near oligo 600-th (red and yellow vertical line), and larger
deletions near 800-th (blue vertical line). On the Figure (b) we present the p-values from K-S tests performed for each oligo
on original design. For each probe three tests were done, which refer to the goodness of fit of oligo in case it is included
in normal, deleted or duplicated segment. These p-values were then used to prepare optimized designs. Figure (c) refers to
the segmentations on the original design. Figures (d), (e), (f) show the results of segmentations performed on the optimized
(reduced) designs. Segmentations on the Figure (d) come from reduced design, that was obtained by uniform removing
random probes from original one. Segmentations on the Figure (e) come from reduced design, that was obtained by uniform
removing most deviated (from segment mean) oligos (lowest p-values from K-S tests). Segmentations on the Figure (f) come
from reduced design, that was obtained by uniform removing least deviated (from segment mean) oligos (highest p-values
from K-S tests).
Summarizing, the robustness measure used in the
optimization context called relative robustness of
smaller array design A with respect to original one
O is defined as follows:
Θ
A|O
=
η
max
∑
η=η
min
σ
tot
η
(7)
ROBUSTNESS OF EXON CGH ARRAY DESIGNS
179

The optimization procedure were preceded by cal-
culation of per-oligo quality score. For each probe in
original design we computed, cumulative properties,
which reflects the oligo suitability in the context of its
surrounding. For a given oligo the K-S tests were per-
formed, which compare the distribution of this oligo
logratio deviations to the distribution of logratio de-
viations taken from the neighborhood of this probe.
The KS-test were performed separately for logratio
assigned to duplicated, deleted and non-aberrated re-
gions. As a result, we obtained three p-values, that
describe the probe functional performance (see Fig-
ure 5b). Those p-values were then used to prepare
optimized designs. Details are presented in Results
Section.
3 RESULTS AND DISCUSSION
3.1 Synthetic Data
Figure 3 presents the comparison of two designs eval-
uated on (synthetic) samples characterized by many
relatively short segments (like in cancer tissues). The
blue color corresponds to good design. Weaker de-
sign (coded in red) contains 20% of poorly hybridiz-
ing probes and 15% of outliers. Hence it corresponds
to generator (d) from the previous Section.
For all oligo probes we present their robustness
Θ
A
i
(upper plot) in logarithmic scale and correspond-
ing importance weights vector ω
A
i
(lower plot). It
is clearly visible, that the robustness is significantly
higher for better (blue) design.
The evaluation of two other designs tested on typ-
ical genomic disorder (not cancer) datasets is illus-
trated in Figure 4. Blue color codes the outcome for
good design and red color corresponds to design con-
taining 15% of poor probes (yielding logratio read-
outs classified as outliers), i. e. datasets from this
design are obtained from generator of type (c). Anal-
ogously as for previous example, the better design
yields higher array robustness.
3.2 Testing Robustness of Optimized
Designs
In previous sections we have shown, that robustness
measure can be useful for estimation of the design
performance in detecting aberrated regions. Below
we present several approaches to aCGH design opti-
mization and the application of robustness in evalua-
tion of those designs quality.
Optimized designs were prepared, based on the
data from 60 aCGH experiments, performed on the
180 K array. The goal was to select 80% of oligos
from original design and keep the ability to detect all
aberrated segments.
Note that our approach operates on different level
of abstraction than those presented in (Xia et al.,
2010) where the probe design factor where calculated.
In our study the research focus is on the functional
performance, i.e., the ability of recovering the real
segmentation.
To investigate the influence of design optimization
strategy on relative array design robustness several
approaches for probes selection were tested, includ-
ing uniform sampling (A
1
design) and most/least suit-
able oligo removal (A
2
and A
3
respectively). Some of
those methods reduced the number of probes with a
little loss of relative robustness. One can benefit from
this strategy especially for targeted arrays used for the
diagnosis of specific chromosomal aberrations.
The comparison shown on the Figure 5 of three
optimized designs to the original one revealed that
segmentations presented on the Figure 5e are the clos-
est to the segmentations on original design - Fig-
ure 5c. Moreover, segmentations on the Figure 5e,
thanks to removing the worst performing probes, de-
tects more aberrations than it is shown on Figure 5c
(see area near oligos 600-th and 800-th).
On the Figure 6 we present the comparison of rel-
ative robustness Θ
A
i
|O
for three different optimized
designs A
i
, i = 1, 2, 3 with respect to the original de-
sign O. On the y-axis the distance σ
tot
η
to the original
segmentation Π is shown, while x-axis presents the
increasing value of noise η.
It is clear that for low values of noise segmen-
tation from optimized and original designs are simi-
lar, which implies the small distance between them.
When the noise is higher, then some of the segments,
that were detected before, disappear. In consequence
the distances between segmentations are growing.
From the Figure 6 we can observe that the de-
sign, obtained by removing most deviated oligos, has
the largest relative robustness (keep the smallest dis-
tance to original segmentation while increasing noise
value).
4 CONCLUSIONS
In this paper we introduced new measures for qual-
ity of CGH array performance. In contrast to previ-
ously proposed approaches we focus on the noise ro-
bustness of segmentation procedure. The method is
tested using appropriately enhanced DNACopy seg-
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
180
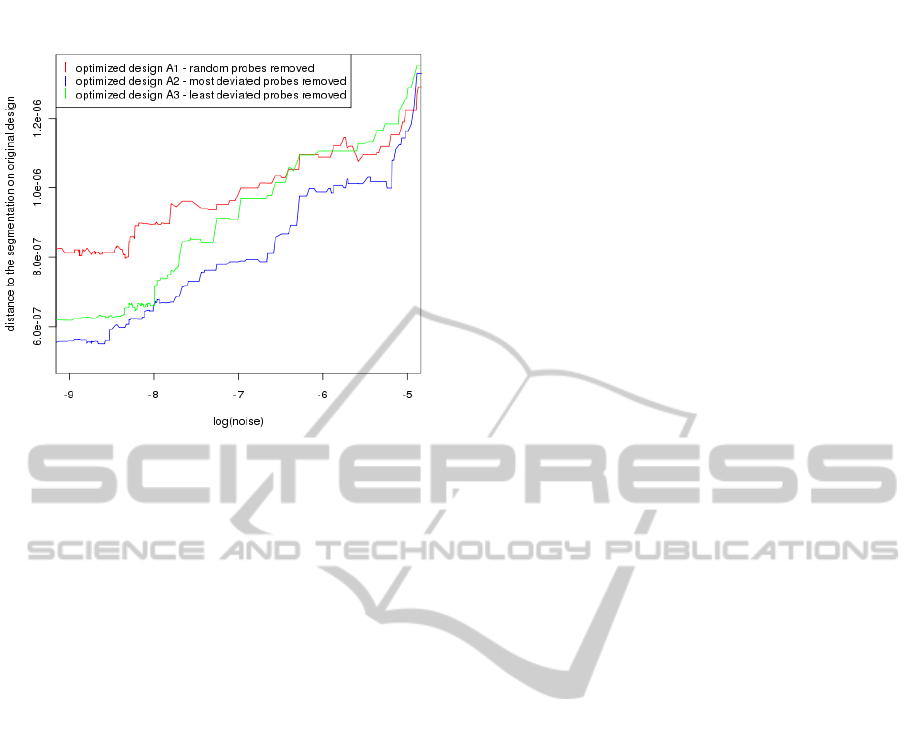
Figure 6: Comparison of relative robustness Θ
A
i
|O
for three
optimized designs.
mentation algorithm (Olshen et al., 2004). Our exper-
iments on real datasets justify the applicability of the
robustness approach. Besides the estimation of the
array performance quality we propose the method to
reduce the array size while keeping its quality on the
reasonable level.
The investigation shows that while optimizing the
design it is crucial to find a tradeoff between keeping
uniform distribution and selecting the best performing
probes. We discovered that the results of design com-
parisons greatly depends on the definition of distance
between two segmentations. Finally, we found new
measure of relative robustness very useful for evalu-
ation of optimized design performance in rearrange-
ments detection.
Several improvements are possible. The chal-
lenging problem is whether DNAcopy segmenta-
tion method may be replaced by more efficient one
(e.g. new segmentation method based on a wavelet
decomposition (Ben-Yaacov and Eldar, 2008)). Also
the noise model used in testing the robustness could
better reflect the real experimental problems.
Authors Contributions. TG, MS and PS designed
the 180 K exon array. TG and MS implemented pro-
grams and carried out the experiments. TG, MS and
AG led the analysis of the experimental results. AG
and PS inspired the robustness approach and super-
vised the project. All authors contributed to the writ-
ing of this manuscript, and have read and approved
the final manuscript.
ACKNOWLEDGEMENTS
This research is supported in part by Polish Ministry
of Science and Educations grants N301 065236, N206
356036 and R13 0005 04. It was also supported by
the Foundation for Polish Science and the European
Social Fund and the State Budget from the Integrated
Regional Operational Program, Action 2.6 ”Regional
Innovation Strategies and Knowledge Transfer”, the
project of Mazovia Voivodship ”Mazovia Doctoral
Scholarship”.
REFERENCES
Baldocchi, R. A., Glynne, R. J., Chin, K., Kowbel, D.,
Collins, C., Mack, D. H., and Gray, J. W. (2005). De-
sign considerations for array CGH to oligonucleotide
arrays. Cytometry. Part A: The Journal of the Inter-
national Society for Analytical Cytology, 67(2):129–
136.
Barrett, M. T., Scheffer, A., Ben-Dor, A., Sampas, N., Lip-
son, D., Kincaid, R., Tsang, P., Curry, B., Baird, K.,
Meltzer, P. S., Yakhini, Z., Bruhn, L., and Laderman,
S. (2004). Comparative genomic hybridization using
oligonucleotide microarrays and total genomic DNA.
Proceedings of the National Academy of Sciences of
the United States of America, 101(51):1776517770.
Ben-Yaacov, E. and Eldar, Y. C. (2008). A fast and flexible
method for the segmentation of aCGH data. Bioinfor-
matics (Oxford, England), 24(16):i139–145.
Cahan, P., Godfrey, L. E., Eis, P. S., Richmond, T. A.,
Selzer, R. R., Brent, M., McLeod, H. L., Ley, T. J.,
and Graubert, T. A. (2008). wuHMM: a robust al-
gorithm to detect DNA copy number variation using
long oligonucleotide microarray data. Nucleic Acids
Research, 36(7):e41.
Carter (2002). Comparative analysis of comparative ge-
nomic hybridization micro array technologies: report
of a workshop sponsored by the wellcome trust. Cy-
tometry, 49(2):43–48.
Caserta, D., Benkhalifa, M., Baldi, M., Fiorentino, F., Qum-
siyeh, M., and Moscarini, M. (2008). Genome pro-
filing of ovarian adenocarcinomas using pangenomic
BACs microarray comparative genomic hybridization.
Molecular Cytogenetics, 1:10.
Chen, H. H., Hsu, F., Jiang, Y., Tsai, M., Yang, P., Meltzer,
P. S., Chuang, E. Y., and Chen, Y. (2008). A probe-
density-based analysis method for array CGH data:
sim ulation, normalization and centralization. Bioin-
formatics (Oxford, England), 24(16):1749–1756.
Coe, B. P., Ylstra, B., Carvalho, B., Meijer, G. A.,
Macaulay, C., and Lam, W. L. (2007). Resolving the
resolution of array CGH. Genomics, 89(5):647–653.
Daz-Uriarte, R. and Rueda, O. M. (2007). ADaCGH: a par-
allelized web-based application and r package for the
analysis of aCGH data. PloS One, 2(1):e737.
ROBUSTNESS OF EXON CGH ARRAY DESIGNS
181

Hampel, F. R., Ronchetti, E. M., Rousseeuw, P. J., and Sta-
hel, W. A. (2005). Robust Statistics: The Approach
Based on Influence Functions. Wiley Series in Proba-
bility and Statistics.
Hijum, S. A. F. T. V., Baerends, R. J. S., Zomer, A. L.,
Karsens, H. A., Martin-Requena, V., Trelles, O., Kok,
J., and Kuipers, O. P. (2008). Supervised lowess
normalization of comparative genome hybridization
data–application to lactococcal strain comparisons.
BMC Bioinformatics, 9:93.
Iourov, I. Y., Vorsanova, S. G., and Yurov, Y. B. (2008).
Chromosomal mosaicism goes global. Molecular Cy-
togenetics, 1:26.
Kreil, D. P. and Russell, R. R. (2005). There is no silver
bullet–a guide to low-level data transforms and nor-
malisation methods for microarray data. Briefings in
Bioinformatics, 6(1):86–97.
Lai, C., Horlings, H. M., de Vijver, M. J. V., Beers, E. H. V.,
Nederlof, P. M., Wessels, L. F., and Reinders, M. J.
(2007). SIRAC: supervised identification of regions
of aberration in aCGH datasets. BMC Bioinformatics,
8:422.
Lemoine, S., Combes, F., and Crom, S. L. (2009). An evalu-
ation of custom microarray applications: the oligonu-
cleotide design challenge. Nucleic Acids Research,
37(6):17261739.
Lipson, D., Aumann, Y., Ben-Dor, A., Linial, N., and
Yakhini, Z. (2006). Efficient calculation of interval
scores for DNA copy number data analysis. Journal of
Computational Biology: A Journal of Computational
Molecular Cell Biology, 13(2):215–228.
Lipson, D., Webb, P., and Yakhini, Z. (2002). Designing
specific oligonucleotide probes for the entire s. cere-
visiae transcriptome. Algorithms in Bioinformatics,
pages 491–505.
Lipson, D., Yakhini, Z., and Aumann, Y. (2007). Optimiza-
tion of probe coverage for high-resolution oligonu-
cleotide acgh. Bioinformatics, 23:e77–83.
Liu, J., Mohammed, J., Carter, J., Ranka, S., Kahveci, T.,
and Baudis, M. (2006). Distance-based clustering of
CGH data. Bioinformatics, 22(16):1971–1978.
Lupski, J. R. (2009). Genomic disorders ten years on.
Genome Medicine, 1(4):42.
O’Hagan, R. C., Brennan, C. W., Strahs, A., Zhang, X.,
Kannan, K., Donovan, M., Cauwels, C., Sharpless,
N. E., Wong, W. H., and Chin, L. (2003). Array
comparative genome hybridization for tumor classifi-
cation and gene discovery in mouse models of malig-
nant melanoma. Cancer Res, 63:5352–5356.
Olshen, A. B., Venkatraman, E. S., Lucito, R., and Wigler,
M. (2004). Circular binary segmentation for the analy-
sis of array-based dna copy number data. Biostatistics
(Oxford, England), 5:557–72.
Perry, G. H., Ben-Dor, A., Tsalenko, A., Sampas, N.,
Rodriguez-Revenga, L., Tran, C. W., Scheffer, A.,
Steinfeld, I., Tsang, P., Yamada, N. A., Park, H. S.,
Kim, J.-I., Seo, J.-S., Yakhini, Z., Laderman, S.,
Bruhn, L., and Lee, C. (2008). The fine-scale and
complex architecture of human copy-number varia-
tion. American journal of human genetics, 82:685–95.
Pollack, J. R., Perou, C. M., Alizadeh, A. A., Eisen, M. B.,
Pergamenschikov, A., Williams, C. F., Jeffrey, S. S.,
Botstein, D., and Brown, P. O. (1999). Genome-wide
analysis of dna copy-number changes using cdna mi-
croarrays. Nature genetics, 23:41–6.
Shaw, C. J., Shaw, C. A., Yu, W., Stankiewicz, P., White,
L. D., Beaudet, A. L., and Lupski, J. R. (2004). Com-
parative genomic hybridisation using a proximal 17p
bac/pac array detects rearrangements responsible for
four genomic disorders. J Med Genet, 41:113–119.
Snijders, A. M., Schmidt, B. L., Fridlyand, J., Dekker, N.,
Pinkel, D., Jordan, R. C. K., and Albertson, D. G.
(2005). Rare amplicons implicate frequent deregula-
tion of cell fate specification pathways in oral squa-
mous cell carcinoma. Oncogene, 24:4232–42.
Staaf, J., Jonsson, G., Ringner, M., and Vallon-Christersson,
J. (2007). Normalization of array-cgh data: influence
of copy number imbalances. BMC Genomics, 8:382.
Thomas, R., Scott, A., Langford, C. F., Fosmire, S. P.,
Jubala, C. M., Lorentzen, T. D., Hitte, C., Karls-
son, E. K., Kirkness, E., Ostrander, E. A., Galibert,
F., Lindblad-Toh, K., Modiano, J. F., and Breen, M.
(2005). Construction of a 2-Mb resolution BAC mi-
croarray for CGH analysis of canine tumors. Genome
Research, 15(12):18311837.
Wang, Y., Makedon, F., and Pearlman, J. (2006). Tumor
classification based on dna copy number aberrations
determined using snp arrays. Oncology reports, 15
Spec no.:1057–9.
Willenbrock, H. and Fridlyand, J. (2005). A comparison
study: applying segmentation to array cgh data for
downstream analyses. Bioinformatics, 21:4084–4091.
Xia, X.-Q., Jia, Z., Porwollik, S., Long, F., Hoemme, C.,
Ye, K., Muller-Tidow, C., McClelland, M., and Wang,
Y. (2010). Evaluating oligonucleotide properties for
DNA microarray probe design. Nucl. Acids Res.,
38(11):e121.
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
182
