
UNIFIED MODELING OF SEVERAL PERTURBATION
EXPERIMENTS IN SYSTEMS BIOLOGY
A Case Study on the Glucose Uptake of Lactococcus Lactis
Andr´as Hartmann
1,∗
, Susana Vinga
1,2
and Jo˜ao M. Lemos
1,3
1
INESC-ID, R. Alves Redol 9, 1000-029 Lisboa, Portugal
2
FCM-UNL, C. M´artires P´atria 130, 1169-056 Lisboa, Portugal
3
IST-UTL, Av. Rovisco Pais, 1049-001 Lisboa, Portugal
Keywords:
Systems biology, Lactococcus lactis, Modeling, Glucose uptake.
Abstract:
Dynamic modeling of the metabolism is one of the main research areas of systems biology. A typical but
yet unresolved problem is the modeling of glucose uptake of Lactococcus lactis bacteria upon in-vivo NMR
measurements in perturbation experiments. Most modelers are focusing on the inverse problem, namely to
identify the parameters of a set of differential equations using the available dataset. Majority of the available
models suffer from the drawback that even if a perfect fit to a single experiment was achieved, they can not
explain the systems’ behavior in different experimental conditions.
The aim of this study is to introduce an appropriate method and a model to fit one set of parameters to several
different experiments, enabling unify modeling of the glucose decay of the bacteria.
With the proposed approach a good overall fit was obtained to the dataset. The results confirm that this could
be a future way towards unified modeling of data with heterogeneous experimental conditions.
1 INTRODUCTION
The aim of inverse modeling is to capture the systems’
dynamic in a set of parameterized Ordinary Differen-
tial Equation (ODE). However, in systems biology the
modeling task is still not certain. Problems can arise
from the available dataset: the distribution of the mea-
surement points might be uneven, and the values are
often manually transcribed. The measurements are in
many cases corrupted with unknown noise, and only
a subset of metabolites are measurable. Other prob-
lems are caused by the the model itself: to capture
the dynamic, nonlinear models are needed, but the
identification of this type of models is far from triv-
ial (Ashyraliyev et al., 2009). Too many parameters
in the model may lead to overparametrisation (Vinga
et al., 2008), sloppyness (Daniels et al., 2008; Vilela
et al., 2009) and / or identifiability problems (Srinath
and Gunawan, 2010), not to mention the need of enor-
mous computationalcapacity because of the exponen-
tially growing parameter space. Finally, many mod-
els are fitted to a single experiment only. Neverthe-
less, the so identified models might explain the given
dataset, but in many cases lack in predictive power
to other experiments. An another consequence of the
above is that even if a very close fit has been achieved
to a single experiment, deterministic models might be
fitted to the unknown noise as well.
The significance of good modeling of the glucose
uptake of Lactococcus lactis bacteria is crucial, be-
cause all the other metabolite concentrations of in-
terest depend on this process, see eg. in (Neves
et al., 2005). Recently, (Castro et al., 2009) revealed
the pathways of glucose uptake, however to our best
knowledge, no unique model fit to different experi-
ments of the glucose uptake has been published before
in the literature. This might be due to the fact that the
sigmoidal shape observed in perturbation experiments
is typically difficult to represent with a simple power-
law function (Goel et al., 2008). Modelers often dis-
regard a good fit to external glucose, as (Goel et al.,
2008), or treat the glucose as input function, eg. (Voit
et al., 2006), even the models that accomplish a good
fit, provide different parameters for different experi-
ments, eg. (Vinga et al., 2008).
Here we introduce an approach based on Particle
Swarm Optimization (PSO) , and a model to glucose
uptake of Lactococcus lactis in perturbation experi-
ments. Our approach has proven useful to fit one set
of model parameters to several different experimental
309
Hartmann A., Vinga S. and M. Lemos J..
UNIFIED MODELING OF SEVERAL PERTURBATION EXPERIMENTS IN SYSTEMS BIOLOGY - A Case Study on the Glucose Uptake of Lactococcus
Lactis.
DOI: 10.5220/0003158103090312
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2011), pages 309-312
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

conditions.
This paper is organized as follows: In section 2
we introduce our method of identification, in section 3
the results are described, and further discussed in sec-
tion 4, finally in section 5 conclusion is drawn and we
point to future work directions.
2 METHODS
In this section first we describe the used dataset, then
the method of identification is introduced, finally we
detail the model of the glucose uptake.
2.1 The Dataset
In vivo Nuclear Magnetic Resonance (NMR) mea-
surements open new horizons for systems biology, al-
lowing measurement of metabolite concentrations in
the living cell (Neves et al., 2005). Unfortunately,
in the case of Lactococcus lactis only the extracel-
lular glucose concentration is measured, and the glu-
cose transport should be modeled. Three perturbation
datasets were used, where a bolus of
13
C labeled glu-
cose was introduced of 20, 40 and 80 mM respectively
to starving bacteria in anaerobe conditions. We also
observed, that the multiple bolus experiments do not
differ much from the single bolus regarding the shape
of the glucose decay.
2.2 Identification Method
Particle Swarm Optimization (PSO) (Kennedy and
Eberhart, 1995) is a population based stochastic op-
timization method inspired by the collective intelli-
gence of simple interacting individuals. The tradi-
tional example for such systems is a bird flock seek-
ing for food. The birds do not know the location of the
food, but their distance from it. Sharing this knowl-
edge with the other members of the flock allows them
to follow the bird that is closest to the food.
In practice, PSO is initialized with a set of possi-
ble solutions, called particles (S
i
) and associated ran-
dom velocities (v
i
). In every iteration (k) the speed
and location of each particle in the parameter space is
updated as
v
i
(k) = wv
i
(k− 1) + c
1
r
1
(pbest
i
− S
i
(k− 1))
+ c
2
r
2
(gbest − S
i
(k− 1)) (1)
S
i
(k) = S
i
(k− 1) + v
i
(k), (2)
where w is the inertia describing the impact of the pre-
vious velocity to the current one. The positive con-
stants c
1
and c
2
correspond to the acceleration rate
towards the local and global optima respectively. r
1
and r
2
are uniform distributed random variables en-
suring the stochastic behavior of the method, pbest
i
is
the best solution discovered so far by the i
th
particle
and gbest is the best solution found in the iteration.
The particle velocities are lower and upper bounded
as v
min
< v
i
< v
max
. The method can be summarized
in the following steps:
1. Initialize a set of particles
2. Evaluate the objective function to all the particles
3. Update gbest and pbest
i
for all particles
4. Count the the new velocities using eq(1)
5. Update the particles’ position using eq (2)
6. Repeat from step 2. until the desired precision or
the limit of iterations is reached.
The objective function here is evaluated in terms of
Mean Squared Errors (MSE) of the fit. This method
was already successfully put into practice for infer-
ring metabolic networks (Naval et al., 2006). Our ap-
proach here is different in the sense that we do not aim
to identify all the metabolites, but only focus on glu-
cose, and instead of fitting to one experiment we use
data of three different experiments (see section 2.1).
2.2.1 Fitting to Multiple Experiments
The trivial way to ensure a fit to multiple datasets is
to use a (weighted) objective function, where the ob-
jective to all the datasets are contributing, for example
by taking the sum of them. The drawback of this ap-
proach is that the summarized error surfaces might be
very complex, and the particles might show very slow
convergence or stuck in a local optima. Thus here we
propose an extension to the method by introducing a
random variables to the objective function as follows
G = q
1
G
1
+ q
2
G
2
+ ... + q
n
G
n
, (3)
where G is the objective function, G
1
...G
n
are the
MSE values according to the datasets 1...n, and
q
1
...q
n
are random values with the following proper-
ties:
∑
n
i=1
q
i
= 1 and E(q
i
) =
1
n
.
2.3 The Model
The idea behind our model is to suppose a variable
(θ), and the glucose concentration (x) depends from it
in a power law manner as shown in eqs (4) and (5).
˙x = −kθ
β
x (4)
˙
θ =
1
c
, (5)
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
310
where k and β are parameters, and c is a constant.
Solving eq (5) results the following simple time de-
pendence: θ = α +
t
c
, introducing a new parameter:
α. To make the model more flexible, we also intro-
duce the constant fraction to the x variable. The final
model is shown in eq (6).
˙x = −k
α+
t
c
β
x
c
(6)
3 RESULTS
First we tested if our model is able to capture the dif-
ferent sigmoidal dynamics derived from the different
glucose uptake time-series. To do this, we applied the
model with the constant c = 1 to the single experi-
ments separately. The model was fitted in 100 inde-
pendent runs of the standard PSO algorithm to each
experiment. The identified parameters and the simu-
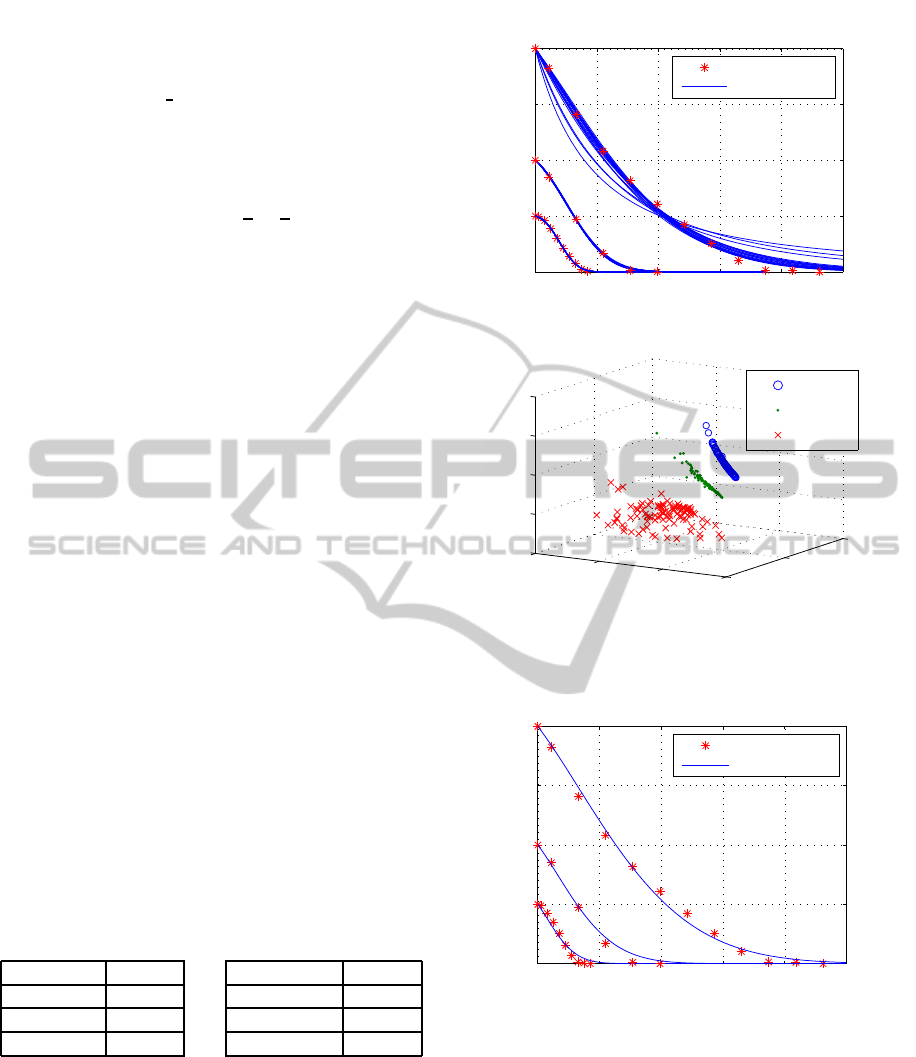
lated time-series are shown in figure 1. As it can be
seen, this approach allows a good identification, how-
ever for the different experimentsthe best revealedpa-
rameters are distinct sets.
For the unified modeling of the different exper-
iments we have chosen c proportional to the initial
glucose pulse as c
20
= 1, c
40
= 2, c
80
= 4 for the 20,
40 and 80 mMol experiment respectively. Here the
extended version of PSO was used for identification
to change the weights dynamically as described in the
previous section. As seen on figure 2 we managed
to achieve an overall good fit to all the three experi-
ments. Using 50 particles, the method discovered the
parameters seen in table 1, here the Mean Squared Er-
rors (MSE) to the experimental data are also shown in
table.
Table 1: The parameters obtained for our model on the three
experiments using the extended version of PSO (left) and
the MSE values to the experiment (right).
Parameter Value
k 0.2631
α 1.0557
β 0.8347
Experiment MSE
20 mM 2.0718
40 mM 2.2654
80 mM 2.9335
4 DISCUSSION
In connection to many models of systems biology it
can be argued that the parameters may not be unique
even within the same experiments. This phenomena
can be interpreted either from the side of the biologi-
cal system as sloppyness, or from the modelers point
of view a poorly constrained model resulting in bad
0 5 10 15 20 25
0
20
40
60
80
Time (min)
Concentration (Mm)
measurements
estimated
0
1
2
0
5
10
15
−1
0
1
2
3
k
α
β
20 mM
40 mM
80 mM
Figure 1: The fit to the time-series (top) stars are the mea-
sured values, solid lines denotes for the estimates via the
100 identified parameter sets on each experiment (bottom).
0 5 10 15 20 25
0
20
40
60
80
Time (min)
Concentration (Mm)
measurements
estimated
Figure 2: The original data points and the estimations with
the best fitting the model.
identifiability. As (Daniels et al., 2008) points out,
different conditions may attract different sets of pa-
rameters of the same biological model. The param-
eters of interest should be chosen from the intersec-
tions of these sets, since using those ensures the good
description of the system within the different condi-
tions. Unfortunately, in the case of using the same
constant c = 1 in the model for different glucose bo-
lus, the explored parameter sets to the single exper-
UNIFIED MODELING OF SEVERAL PERTURBATION EXPERIMENTS IN SYSTEMS BIOLOGY - A Case Study on
the Glucose Uptake of Lactococcus Lactis
311

iments do not overlap, see figure 1. In our interpre-
tation this indicates the inadequacy of this approach.
However the model fits remarkably to individual ex-
periments, we would like to point out that this only
proves the models flexibility to capture the different
sigmoidal shapes.
By introducing a constant to the model, propor-
tional to the initial glucose bolus we managed to
achieve a good overall fit of the model with the same
set of parameters on different experiments (figure 2).
The trade-off is that MSE values to particular exper-
iments are moderately high. We found that our ap-
proach was adequate in fitting several experiments.
The random variables in the objective function re-
sulted that the algorithm is dynamically changing the
weights between the experiments, and ensures a good
convergence even if the sum of the error surfaces
would get difficult.
The sigmoid shape of the glucose uptake was found
slightly varying on the different experiments. This
might be a consequence of the different activity of
the glucose uptake systems revealed by (Castro et al.,
2009), or the differences between the transport of the
glucose monomers. We are also aware of that the glu-
cose uptake might be influenced by other factors, for
example the biomass, a feedback mechanism from the
inside of the cell or the energy level of the cell accord-
ing to (Papagianni et al., 2007). The model could be
extended to involve these aspects.
5 CONCLUSIONS AND FUTURE
WORK
Here we introduced a model to Lactococcus lactis
glucose uptake, and an approach based on PSO to fit it
to three glucose perturbation experiments with differ-
ent glucose input. With our approach a good overall
fit was achieved to the data using one set of parame-
ters. We think that this could be a future way towards
unified modeling of data with different experimental
conditions.
We can not exclude that our model is not complete,
and additional terms might be missing from it. Our
future work will aim to identify these terms. We
are also considering the distinct modeling of the glu-
cose monomers and to extend the identification of the
model to aerobic conditions.
ACKNOWLEDGEMENTS
This work was supported by project DynaMo
(PTDC/EEA-ACR/69530/2006) FCT, Portugal.
REFERENCES
Ashyraliyev, M., Fomekong-Nanfack, Y., Kaandorp, J. a.,
and Blom, J. G. (2009). Systems biology: parameter
estimation for biochemical models. The FEBS jour-
nal, 276(4):886–902.
Castro, R., Neves, A. R., Fonseca, L. L., Pool, W. a., Kok,
J., Kuipers, O. P., and Santos, H. (2009). Charac-
terization of the individual glucose uptake systems
of Lactococcus lactis: mannose-PTS, cellobiose-PTS
and the novel GlcU permease. Molecular microbiol-
ogy, 71(3):795–806.
Daniels, B. C., Chen, Y.-J., Sethna, J. P., Gutenkunst, R. N.,
and Myers, C. R. (2008). Sloppiness, robustness, and
evolvability in systems biology. Current opinion in
biotechnology, 19(4):389–95.
Goel, G., Chou, I.-C., and Voit, E. (2008). System estima-
tion from metabolic time-series data. Bioinformatics,
24(21):2505.
Kennedy, J. and Eberhart, R. (1995). Particle swarm opti-
mization. of IEEE international conference on neural,
4:1942–1948.
Naval, P., Sison, L., and Mendoza, E. (2006). Metabolic
network parameter inference using particle swarm op-
timization. In International Conference on Molecular
Systems Biology, pages 3–4.
Neves, A. R., Pool, W. a., Kok, J., Kuipers, O. P., and San-
tos, H. (2005). Overview on sugar metabolism and its
control in Lactococcus lactis - the input from in vivo
NMR. FEMS microbiology reviews, 29(3):531–54.
Papagianni, M., Avramidis, N., and Filiousis, G. (2007).
Glycolysis and the regulation of glucose transport in
Lactococcus lactis spp. lactis in batch and fed-batch
culture. Microbial cell factories, 6:16.
Srinath, S. and Gunawan, R. (2010). Parameter identifiabil-
ity of power-law biochemical system models. Journal
of biotechnology.
Vilela, M., Vinga, S., Maia, M. a. G. M., Voit, E. O., and
Almeida, J. S. (2009). Identification of neutral bio-
chemical network models from time series data. BMC
systems biology, 3:47.
Vinga, S., Thomaseth, K., Lemos, J., Neves, A., Santos,
H., and Freitas, A. (2008). Structural analysis of
metabolic networks: a case study on lactococcus lac-
tis. In 8th Portuguese Conference on Automatic Con-
trol (CONTROLO2008), pages 566–571.
Voit, E., Neves, A., and Santos, H. (2006). The intricate
side of systems biology. Proceedings of the National
Academy of Sciences, 103(25):9452.
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
312
