
NEPs-Lingua: A New Textual Language to Program
NEPs
M. de la Cruz
1
, A. Jim´enez
1
, E. del Rosal
1
, G. Bel-Enguix
2
and A. Ortega
1
1
Departamento de Ingenier´ıa Inform´atica, Universidad Aut´onoma de Madrid, Madrid, Spain
2
GRLMC-Research Group on Mathematical Linguistics, Universitat Rovira i Virgili
Tarragona, Spain
Abstract.
1
Networks of Evolutionary Processors (NEPs) are one of the currently
most used new types of natural computers. This paper briefly describes the model
and some developing tools for NEPs. Then, it describes NEPs-Lingua, a new
textual programming language for NEPs. Its two main goals are: reducing the size
needed by other representations and keeping the syntax as close as possible to the
one used to define NEPs in the literature. Some examples and future research
lines are also discussed.
1 Motivation
A great deal of research effort is currently being made in the so called “natural comput-
ing” realm. “Natural computing” is mainly focused on the definition, formal descrip-
tion, analysis, simulation and programmingof newmodels of computation (usually with
the same expressive power as Turing Machines) inspired by Nature. Their bio-inspired
nature makes these models specially suitable for the simulation of complex systems.
Some of the best known natural computers are Lindenmayer systems (L-systems, a
kind of grammars with parallel derivation), cellular automata, DNA computing, genetic
and evolutionary algorithms, multi agent systems, artificial neural networks, P-systems
(computation inspired by membranes) and NEPs (or networks of evolutionary proces-
sors). This paper is devoted to this last model.
There are two main areas in which these models could be useful: as new architec-
tures for computers, different from von Neumann’s machine; and as modelling tools
to simulate complex systems for which “conventional approaches” (usually based on
differential equations) are, in practice, difficult to handle.
Two steps are needed in both scenarios: 1) design a particular instance of the consid-
ered model able to solve the task under study (this step is equivalent to “programming”
the model) and 2) “run” the model.
1
This work was partially supported by the R&D program of the Community of Madrid
(S2009/TIC-1650, project “e-Madrid”) as well as by MTM 2007-63422. The authors thank
Dr. Manuel Alfonseca for his help to prepare this document.
de la Cruz M., Jiménez A., del Rosal E., Bel-Enguix G. and Ortega A..
NEPs-Lingua: A New Textual Language to Program NEPs.
DOI: 10.5220/0003308200370046
In Proceedings of the 1st International Workshop on AI Methods for Interdisciplinary Research in Language and Biology (BILC-2011), pages 37-46
ISBN: 978-989-8425-42-3
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

Several attemps have been made to build hardware devices to support these bio-
inspired models. Some research groups are currently implementing in silico the basic
components of P-systems [4]. [9] describes other examples of hardware implementa-
tions of cellular automata, CAM-6 and its derivatives, that have been used for the simu-
lation of complex systems (see [8]). But, unfortunately there are no real computers for
almost all bio-inspired models. So, step 2 usually implies the simulation of the model
in a “conventional” (von Neumann) computer.
Informally, and assuming that NP (nondeterministic polynomial time) 6= P, NP is a
complexity class that includes those problems whose solution by means of algorithms
run on conventional computers requires more than polynomial time. We can informally
understand more than polynomial as exponential. One of the most interesting features
of these bio-inspired computers is their intrinsic parallelism. We can design algorithms
for them that could improve the exponential performance of their classic versions. Nev-
ertheless, when the models have to be simulated on conventional computers, the total
amount of space needed to simulate the model and to actually run the algorithm usually
becomes exponential. This may be one of the main reasons why natural computers are
not widely used to solve real problems. Most of the simulators are not able to handle
the size of non trivial problems. Grid, cloud computation and clusters offer an inter-
esting and promising option to overcome the drawbacks of both solutions: “specific”
hardware, and simulators run on von Neumann’s machines.
There are several research groups interested in programming tools for natural com-
puters. These tools include textual and visual programming languages, compilers, se-
quential and parallel simulators.
P-Lingua ([5] and http://www.p-lingua.org) is a programming language for
membrane computing which aims to be a standard to define P systems. One of its main
characteristics is to remain as close as possible to the formal notation used in the liter-
ature to define P systems. Once he has formalized his P systems, the programmer does
not need any additional effort to describe them with P-Lingua. P-Lingua is also the
name of a software package that includes several built-in simulators for each supported
model, as well as the compilers needed to simulate P-Lingua programs.
One of the current topics of interest of the authors of this paper is the development of
programming tools for NEPs, which will be briefly described in the following sections.
The current paper introduces NEPs-Lingua, the first textual programming language for
NEPs. It is a first step to extend the P-Lingua approach to other bio-inspired models
of computation. Our goal is to provide the researchers with a homogeneous family of
languages for programming natural computers. The programmer familiar with a model
will not have to learn a very different syntax if he tries to use other models. This is the
reason why NEPs-Lingua is designed to be similar to P-Lingua. NEps-Lingua has two
main goals that will also be described in detail later: 1) Like P-Lingua, it aims to provide
the researchers with a syntax as close as possible to the one used to describe NEPs in
the literature. 2) It tries to ease some usually boring, mechanical and time-consuming
tasks needed to describe NEPs with the input formalisms of the available tools.
38

2 NEPs
A Network of evolutionary processors (NEP [1]) can be defined as a graph whose nodes
are processors which perform very simple operations on strings and send the resulting
strings to other nodes. Every node has filters that block some strings from being sent
and/or received.
NEPs-Lingua also support PNEPs (Parsing NEPs), an extension to NEPs introduced
in [7] to handle context free grammars.
NEPs and PNEPs: Definitions and Key Features. Following [1] we introduce the
basic definition of NEPs.
Definition A Network of Evolutionary Processors of size n is a construct:
Γ = (V, N
1
, N
2
, ..., N
n
, G),
where:
– V is an alphabet and for each i with 1 ≤ i ≤ n,
– N
i
= (M
i
, A
i
, P I
i
, P O
i
) is the i-th evolutionary node processor of the network.
The parameters of every processor are:
• M
i
is a finite set of evolution rules of just one of the following forms:
i. a → b, where a, b ∈ V (substitution rules),
ii. a → ε, where a ∈ V (deletion rules),
iii. ε → a, where a ∈ V (insertion rules),
iv. a → s, where a ∈ V , s ∈ V
∗
(context free rules applied to change a
symbol by a string) PNEPs replace substitution rules by this kind of rules.
PNEPs add this kind of rule to reduce the amount of equivalentderivations.
• A
i
is a finite set of strings over V . The set A
i
is the set of initial strings in the
i-th node.
• P I
i
and P O
i
respectively representing the input and the output filters. These
filters are defined by the membership condition, namely a string w ∈ V
∗
can
pass the input filter (the output filter) if w ∈ P I
i
(w ∈ P O
i
). In this paper we
will use two kind of filters:
∗ Those defined as two components (P, F ) of Permitting and Forbidding
contexts (a word w passes the filter if ( alphabet of w ⊆ P )∧(F ∩ alphabet
of w = ∅)).
∗ Those defined as regular expressions r (a word w passes the filter if w ∈
L(r), where L(r) is the language defined by the regular expression r).
– G = ({N
1
, N
2
, . . . , N
n
}, E) is an undirected graph called the underlying graph of
the network. The edges of G, that is the elements of E, are given in the form of sets
of two nodes. The complete graph with n vertices is denoted by K
n
.
Further formal details on the way in which NEPs evolve can be found in [1].
39

3 Programming Tools for NEPs
The authors of this paper have proposed a development environment for programming
NEPs that includes a Java NEP simulator (jNEP), a Java graphical viewer of its sim-
ulations (jNEPview), and a domain specific visual language for NEPs designed with
AToM
3
(NEPVL) In the following paragraphs we will briefly introduce these modules.
NEPs-Lingua and its compilers will be integrated in this environment as its textual pro-
gramming language.
jNEP and jNEPview. jNEP [3] reads the definition of the NEP from an XML configu-
ration file that contains special tags for any relevant components in the NEP (alphabet,
stopping conditions, the complete graph, every edge, the evolutionary processors with
their respective rules, filters and initial contents). Despite the complexity of these XML
files, the interested reader can see that the tags and their attributes have self-explaining
names and values.
We show below, as an example, the configuration file of a very simple NEP. It has
two nodes that, respectively,delete and insert the symbol B. The initial word AB travels
from one node to the other. The first node removes the symbol B from the string before
leaving it in the net. The other node receives the string A and adds again the symbol
B. The resulting string comes back to the initial node and the same process takes place
again.
<NEP nodes="2">
<ALPHABET symbols="A_B"/>
<GRAPH> <EDGE vertex1="0" vertex2="1"/> </GRAPH>
<EVOLUTIONARY_PROCESSORS>
<NODE initCond="A_B">
<EVOLUTIONARY_RULES>
<RULE ruleType="deletion" actionType="RIGHT" symbol="B"
newSymbol=""/></EVOLUTIONARY_RULES>
<FILTERS> <INPUT type="2" permittingContext="A_B"
forbiddingContext=""/>
<OUTPUT type="2" permittingContext="A_B"
forbiddingContext=""/></FILTERS>
</NODE>
<NODE initCond="">
<EVOLUTIONARY_RULES>
<RULE ruleType="insertion" actionType="RIGHT" symbol="B"
newSymbol=""/> </EVOLUTIONARY_RULES>
<FILTERS> <INPUT type="2" permittingContext="A_B"
forbiddingContext=""/>
<OUTPUT type="2" permittingContext="A_B"
forbiddingContext=""/></FILTERS>
</NODE>
</EVOLUTIONARY_PROCESSORS>
<STOPPING_CONDITION>
<CONDITION type="MaximumStepsStoppingCondition" maximum="8"/>
</STOPPING_CONDITION>
</NEP>
(XML configuration file for a simple NEP with just two processors that send the words A and B
back and forth)
40

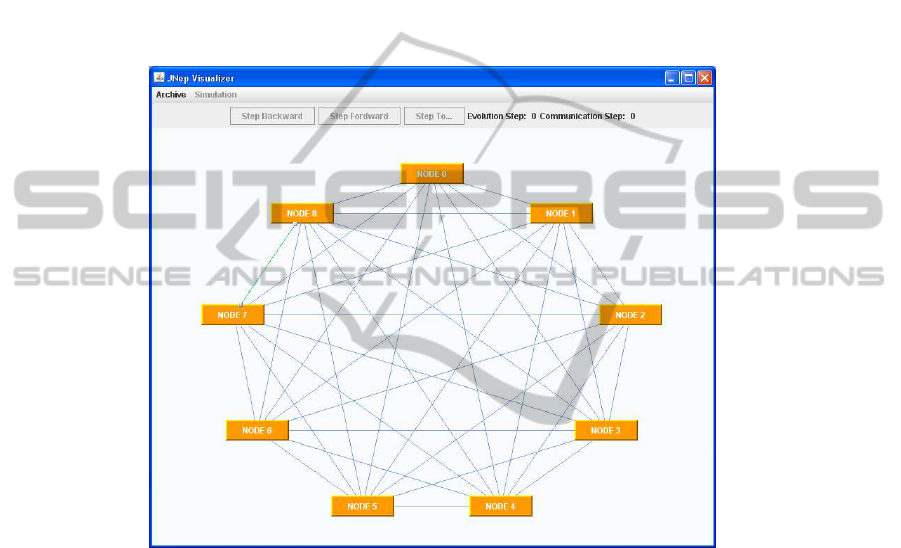
Figure 2 contains, as an example, the windows used by jNEPview to show the net-
work topology of one of the NEPs under study. Further details on jNEPview can be
found in [2].
NEPVL. AToM
3
is a python platform to develop domain specific visual languages. We
have used it to design NEPVL.
Fig.1. NEPVL program for the NEP with two processors.
Figure 1 shows the NEPVL program that defines the same NEP with two processors
previously described. Further details about AToM
3
and NEPVL can be found in [6].
4 The NEPs-Lingua Syntax
In the following paragraphs we describe, mainly by examples, the syntax of NEPs-
Lingua. A full ANTLR
2
description of the complete grammar may be asked from the
authors. The main components of a NEPs-Lingua program are atomic data, comments,
nodes, the alphabet, the initial contents of the nodes, evolutionary rules, filters, the
connections of the NEP graph and stopping conditions.
Atoms. There are two classes of atomic data: alfanumeric strings of symbols (they have
to start with an alphabetic character); and integer arithmetic expressions, with the usual
mathematical notation, that include the operators in the set {∧(power), +, −, ∗, /}
Comments. The typical C++ comments are also available in NEPs-Lingua.
2
ANTLR is a Java tool to design top-down parsers and language processors, developed by
Terence Par. Further information can be found at http://www.antlr.org/
41

Line Comments. For example // Comment.
The comment includes every symbol until the end of the line.
Multi Line comments. For example
/
*
... Comment
...
*
/
Where the comment includes everything (even the end of line markers) between the
symbols “/*” and “*/”.
Alphabet. It is the alphabet of the NEP, a set of strings of symbols. The expression
@A={X,S,a,b,o,O} defines an alphabet that contains the elements “X”, “S”, “a”, “b”,
“O”, and “o”.
Nodes. This is the most complex type of NEPs-Lingua data. There are two classes of
nodes: with and without indexes. There are two kinds of indexes: numeric (defined by
a range) and symbolic (defined by a set of strings of symbols). The syntax of indexes
with numeric ranges is borrowed from P-Lingua.
Non Indexed Nodes. The expression {initial, final} defines two nodes without
indexes with names initial and final.
Indexed Nodes. The example defines a family of nodes with two indexes. One of them
(i) takes its values from the interval [0, 10]. The values of the other (j) are taken from
the set {o, a, b}.
{m{i,j}: 0<=i<=10, j->{o,a,b}}
The explicit set of the 33 defined nodes is {m
0,a
, m
0,b
, m
0,c
, . . . m
10,a
, m
10,b
, m
10,c
}.
Different kinds of nodes can be mixed by means of the union operator. The next
example shows the definition of a set of nodes that contains the two previous examples.
@N={initial, final}+{m{i,j}: 0<=i<=10, j->{o,a,b}}
Initial Content. It describes the set of strings that a given node initially contains.
Notice that the node is written as a parameter of the content directive @c. The expression
@c{n{X}} = {X, S} sets the initial content of the node n
X
to {X, S}
Rules. Each type of rule has a different notation. Notice that, as in P-Lingua, the symbol
# stands for the empty string and the string --> separates the left and right sides of
the rule. The sentences # -->a, a --># and S-->aSb are examples of respectively
insertion, deletion, and substitution (or deriving) rules.
All the rules for a given node are given together in the same sentence. The sentence
@r{n{S}} = {S-->aSb, S-->ab} assigns two deriving rules to the node n
S
.
42

Filters. Each processor needs an input and an output filter. Different papers previously
mentioned define three components in the filters: their type and the permitting and for-
bidding contexts. We have grouped the different filters of the literature in six types
(depending on the way in which they are applied): types from 1 to 4 and filters defined
by means of regular expressions or by means of sets of strings. Both contexts are just
sets of symbols described by means of regular patterns or explicit sets of strings. The
following examples define several filters:
@pif{n{S}}= {1, {abc, oo}}
@fof{initial} = {@regular_pattern, ( ((a[]b)+) ][ (c
*
) )][ # }
@pif{n{2,a}}= {@set, {a,ab,aabb}}
where @pif and @fof stand respectively for permitting input and forbidding ouput
filter (the same for forbiddinginput and permitting output filters). In regular expressions
[], ][, +,
*
, # represent intersection, union, + and *, and the empty string.
Connections. This element makes it possible to get a compact representation of NEPs.
There are two ways of defining connections: the directive @complete, that stands for a
complete graph; and an explicit set of connections defined by means of pairs of nodes.
The next examples show both options:
@C=@complete
@C={ (final,n{X}), (n{X},m{9,a}) }
Stopping conditions. The stopping conditions are written in a set after the directive
@S. Each kind of condition is represented by its name and its required parameters. Both
names and parameters are easy to identify in the following example:
@S={@no_change, @max_steps = 3+4, @non_emtpy_node={n{O}, n{X}} }
where @no_change stands for two consecutive equal configurations; @max_steps re-
quires an expression to define the number of steps (the NEP stops after taking the given
number of steps); and @non_empty_node includes a set of nodes whose contents are
initially empty (the NEP stops when one of these nodes receives some string).
5 Examples
In this section we will show some complete NEPs-Lingua programs. Our main goal is to
highlight the two main characteristics of NEPs-Lingua: reducing the size and keeping
close to the formal notation. For this purpose we will compare several NEPs-Lingua
programs with NEPs examples taken from the literature. For space reasons, we refer to
the original papers for the detailed definition of the examples.
Reducing the Size of the Representations. First we show the NEPs-Lingua program
for the example with two processors previously described. We can appreciate with this
simple example that the NEPs-Lingua program is more compact than the other two
representations (see the corresponding XML source and figure 1).
43
@A={A,B}
@N={ n{i}: 0 <= i <= 1}
@c{n{0}}={A,B}
@r{n{0}}={B-->#}
@r{n{1}}={#-->B}
@S={@max_steps = 8 }
@C={@complete}
The reduction in size is greater as the complexity of the NEP increases. NEPs usu-
ally have complete graphs.
Fig.2. NEPVL program for the NEP that solves the 3 SAT problem.
Figure 2 shows the jNEPview window for a NEP with a complete graph with 9
nodes.
The XML configuration file for this NEP is forced to explicitly contain all the
nodes and connections(see the XML sample previously shown) while the NEPs-Lingua
source has to contain just the following two sentences:
@N={ n{i}: 0 <= i <= 8}
@C=@complete
[1] shows a NEP able to solve a small instance of the well known graph coloring
problem with three different colours. It needs a complete graph with more many nodes
than in the previous example.
44

The jNEPview window for this NEP is not shown in this paper because it is difficult
to handle: it looks like a ball of yarn. Once again the NEPs-Lingua program needs just
the following two sentences:
@N={ n{i}: 0 <= i <= 50 } \\ Definition of 51 nodes
@C=@complete
Keeping NEPs-Lingua as Close as Possible to the Formal Notation used in the Lit-
erature. The interested reader can easily see in the references for the last two examples
(3-SAT and 3 coloring) that NEPs-Lingua syntax is mainly inspired by the formal no-
tation used in the literature to describe NEPs.
[7] contains another example: a NEP associated with the context free grammar for
axiom X with the derivation rules {X → SO, S → aSb, S → ab, O → o, O →
oO, O → Oo}
It is easy to see that the following NEPs-Lingua program for this NEP is quite
similar to its formal definition.
@A={X,S,a,b,o,O} // Alphabet
@N= {final}+ {n{symbol}:symbol->{X,S,O}} /
*
Nodes associated
with non terminal symbols
*
/
@c{n{X}}={X} // Initial content of the axiom node
@r{n{X}}= {X-->SO} // Deriving rules for the axiom
@r{n{S}}= {S-->aSb, S-->ab}
@r{n{O}}= {O-->o, O-->oO, O-->Oo}
@C=@complete // The graph is complete
@S={ @non_emtpy_node={final} } // Stopping conditions
6 NEPs Lingua Semantics
The semantic constraints that every NEPs-Lingua program has to satisfy are outlined
below:
– It has to contain exactly one alphabet and one set of node declarations.
– It needs at most one of the following elements:
• Connection declaration set. By default, the graph is considered complete.
• Set of stopping conditions. @no_change is assumed by default.
– Filters, rules and initial contents are optional.
– Nodes have to be defined before their use.
– Each symbol representing rules, filters and initial contents has to be included in the
alphabet.
NEPs-Lingua compilers should ensure these conditions. The usual way of control-
ling the last one is by means of a symbol table that is filled while processing the decla-
ration sentences and is consulted by the sentences that use nodes and symbols.
We have used different Hashtable Java objects to check these constrains.
The following example shows some semantic mistakes:
45

@A={A}
@N={ n{i}: 0 <= j <= 1}
@c{n{0}}={A,B}
@r{n{0}}={B-->#}
@r{n{2}}={#-->B}
@S={@max_steps = 8 }
@C={@complete}
– The third, fourth and fifth lines contain the symbol B, which is not in the alphabet.
– The second line defines the index j, while the declared one is i
– The fifth line defines the rules for the node n
2
, but the value for index (2) is invalid
7 Further Research Lines
Code Generators. In the future we intend to provide our system with some code gen-
erators. The first one will translate NEPs-Lingua programs into the XML configuration
files that jNEP uses as input. It could also be used to generate the same python programs
that the NEPVL software uses.
Extensions. Although this simple syntax seems to be expressive enough for the NEPs
we have found in the literature, we are considering the following extensions, usually
present in programming languages: global variables and parametrized sub-NEPs.
References
1. J. Castellanos, C. Mart´ın-Vide, V. Mitrana, and J. M. Sempere. Networks of evolutionary
processors. Acta Informatica, 39(6-7):517–529, 2003.
2. M. Cu´ellar and E. del Rosal. jnepview: a graphical trace viewer for the simulations of neps.
3. E. del Rosal, R. Nu˜nez, C. Casta˜neda, and A. Ortega. Simulating neps in a cluster with jnep.
In Proceedings of International Conference on Computers, Communications and Control, IC-
CCC 2008,, 2008.
4. L. Fern´andez, V. J. Mart´ınez, and L. F. Mingo. A hardware circuit for selecting active rules in
transition p systems. In Seventh International Symposium on Symbolic and Numeric Algo-
rithms for Scientific Computing (SYNASC 2005), 2005.
5. M. Garc´ıa-Quismondo, R. Guti´errez-Escudero, M. A. Mart´ınez del Amor, E. Orejuela, and
I. P´erez-Hurtado. P-lingua 2.0: A software framework for cell–like p systems. International
Journal of Computers, Communications and Control, IV(3):234–243, 2009.
6. A. Jim´enez, E. del Rosal, and J. de Lara. A visual language for modelling and simulation
of networks of evolutionary processors. In Trends in Practical Applications of Agents and
Multiagent Systems, Advances in Soft Computing.
7. A. Ortega, E. del Rosal, Diana P´erez, R. Mercas¸, A. Perekrestenko, and M. Alfonseca. PNEPs,
NEPs for Context Free Parsing: Application to Natural Language Processing, pages 472–479.
LNCS. 2009.
8. M. A. Smith and Y. Bar-Yam. Cellular automaton simulation of pulsed field gel electrophore-
sis. ELECTROPHORESIS, 14(1):1522–2683, 1993.
9. T. Toffoli and N. Margolus. Cellular Automata Machines. MIT Press, London, 1987.
46
