
A WEB-BASED REPOSITORY OF REPRODUCIBLE SIMULATION
EXPERIMENTS FOR SYSTEMS BIOLOGY
Michael A. Guravage and Roeland M. H. Merks
Centrum Wiskunde & Informatica (CWI)
Netherlands Consortium for Systems Biology & Netherlands Institute for Systems Biology (NCSB-NISB)
Science Park 123, 1098 XG Amsterdam, The Netherlands
Keywords:
SED-ML, SBML, MIASE, Simulation, Modeling, Plone, CMS.
Abstract:
Systems Biology requires increasingly complex simulation models. Effectively interpreting and building upon
previous simulation results is both difficult and time consuming. Thus, simulation results often cannot be
reproduced exactly; making it difficult for other modellers to validate results and take the next step in a
simulation study.
The Simulation Experiment Description Mark-up Language (SED-ML), a subset of the Minimum Information
About a Simulation Experiment (MIASE) guidelines, promises to solve this problem by prescribing the form
and content of the information required to reproduce simulation experiments. SED-ML is detailed enough to
enable automatic rerunning of simulation experiments.
Here, we present a web-based simulation-experiment repository that lets modellers develop SED-ML compli-
ant simulation-experiment descriptions The system encourages modellers to annotate their experiments with
text and images, experimental data and domain meta-information. These informal annotations aid organi-
sation and classification of the simulations and provide rich search criteria. They complement SED-ML’s
formal precision to produce simulation-experiment descriptions that can be understood by both men and ma-
chines. The system combines both human-readable and formal machine-readable content, thus ensuring exact
reproducibility of the simulation results of a modelling study.
1 INTRODUCTION
Systems biologists unravel the inner workings of bi-
ological systems in a continuous cross-talk between
predictive computer simulation models, and biolog-
ical experiments to test the model predictions. To
build models efficiently and compare model predic-
tions with experiments, effectively interpreting sim-
ulation results and building upon previous work is
crucial. Unfortunately, it is both difficult and time
consuming, because, 1) publishing exact model def-
initions and working simulation codes is not (yet)
common practice, and 2) the exact steps taken and
the information required for reproducing the simula-
tion (e.g. model parameters) are rarely recorded ex-
actly. As a result, trying to reproduce published sim-
ulation results can become a research project in itself.
This hampers progress in systems biology. The dif-
ficulty in validating results impedes the progress of
simulation studies.
A range of public databases exists for depositing
computational models in a systematic way, including
the BioModels database (Le Nov
`
ere et al., 2006), the
modelDB (Hines et al., 2004), the Database of Quan-
titative Cellular Signalling (DOQCS) (Sivakumaran
et al., 2003) and Physiome (Yu et al., 2009; Yu et al.,
2011). Although these databases make it much easier
to retrieve the definitions and codes of published sim-
ulation models, reproducing an observation in a sim-
ulation model also requires exact knowledge of many
additional factors including the initial conditions, the
parameters used, the integration times, etc. In other
words, the model databases contain the ingredients
for a simulation experiment, but the recipe is lacking.
The Simulation Experiment Description Markup
Language (SED-ML) (K
¨
ohn and Le Nov
`
ere, 2008)
implements a subset of the Minimum Information
About a Simulation Experiment (MIASE) guidelines.
MIASE is an emerging standard that promises to
solve this problem by prescribing the form and con-
tent of the information required to reproduce simula-
tion experiments. SED-ML is a recipe template, an
instance of which include a description of and refer-
ence to existing, preferably curated, models, inputs,
134
A. Guravage M. and M. H. Merks R..
A WEB-BASED REPOSITORY OF REPRODUCIBLE SIMULATION EXPERIMENTS FOR SYSTEMS BIOLOGY.
DOI: 10.5220/0003598001340141
In Proceedings of 1st International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH-2011), pages
134-141
ISBN: 978-989-8425-78-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

procedures and outputs that, together, describe of how
to use the models in simulations that constitute an ex-
periment. While SED-ML is detailed enough to en-
able automatic rerunning of simulation experiments,
its XML-based format makes it difficult for modellers
to interpret. Also, SED-ML focuses on computational
models of biological processes written in, e.g., SBML
format; whereas the simulations most difficult to re-
produce are typically written in general-purpose lan-
guages. Our repository accommodates models writ-
ten in both XML and general-purpose languages like
C++ by allowing model references to include both
model databases and source code repositories.
SED-ML is an grammar for describing simulation
experiments. Its specificity allows one to describe a
simulation experiment in sufficient detail to reproduce
the simulation results automatically. SED-ML even-
tually can act as a scripting language to describe the
steps leading to a published simulation result. While
SED-ML ensures that the simulation-experiment de-
scription is unambiguous, it is difficult to interpret
the intention of the modeller by reading only his
SED-ML code. Combining complimentary informal
descriptive information with SED-ML’s formal spec-
ification ensures that simulation experiments descrip-
tions can be interpreted by both humans and ma-
chines. Adherents of Literate Programming (Knuth,
1984) have long argued that the complementary na-
ture of informal descriptions and formal specifica-
tions facilitates the retention and transfer of informa-
tion.
For the modeller, creating simulation-experiment
descriptions in our repository is analogous to keep-
ing a laboratory notebook. Models can be described
in the context of real simulations at an appropriate
level of abstraction and detail, and the complementary
nature of informal and formal specifications encour-
ages correctness and completeness. As a result, sim-
ulation experiments are both more easily reproduced
and more readily understood. Using the repository
thus facilitates collaboration between modellers. The
repository can be tailored to realise any collaboration
strategy, e.g., a repository’s contents can be accessi-
ble to the public or restricted to an enumerated list of
individuals. Modelers can invite colleagues to col-
laborate on specific simulation-experiment descrip-
tions. Workflows shepherd simulation-experiment de-
scriptions through a series of states from inception
to completion; so a modeller can easily see which
simulation-experiment descriptions are in develop-
ment, which have been submitted for approval and
which have been tested and validated. Experimen-
tal collaborators can evaluate a repository of quali-
fied simulation-experiment descriptions that describe
models in the context of a real simulation.
In this paper, we argue that placing simulation-
experiment descriptions in a repository that allows an-
notating them with prose descriptions, domain meta-
information and experimental data, and that guides
their progress through a publication workflow, will
significantly improve the usability of the SED-ML
recipes. Our final goal is to see published simula-
tion experiments refer to simulation-experiment de-
scriptions in our repository. Those descriptions will
contain both machine-executable and human-readable
recipes that allow modellers to interpret, evaluate and
reproduce the simulations and build upon the pub-
lished results.
2 THE MODEL REPOSITORY
2.1 Requirements
We aimed to construct a system that allows mod-
ellers to create simulation-experiment descriptions
conforming to the MIASE guidelines, according to
the following system requirements:
2.1.1 Ease of Use, Encourage Annotation
The system should help modellers along the process
of creating and managing their simulation-experiment
in ways that are natural and easy for them. The top
level of a simulation-experiment description hierar-
chy is comprised of five classes:
• Models
• Simulations
• Tasks
• Outputs
• Data Generators
The system assists the modeller in creating the
various elements in the correct order and with valid
data. In addition to the minimum data prescribed by
the MIASE guidelines, the system should encourage
the modeller to annotate each part of a simulation-
experiment description with descriptive information
at an appropriate level of abstraction. The editing
environment should be familiar to anyone who has
worked with office suite software. The system should
also allow the user to include experimental data such
as images and animations, and generic and domain-
specific meta-information. This data should be in-
dexed automatically and be made available as search
criteria.
A WEB-BASED REPOSITORY OF REPRODUCIBLE SIMULATION EXPERIMENTS FOR SYSTEMS BIOLOGY
135

2.1.2 Data Security
The system should have several levels of security,
such that users can choose to keep a project from
themselves, share it with coworkers or reviewers,
or make it public after the results have been pub-
lished in a scientific journal. Modelers own their
simulation-experiment description, and they cannot
be viewed or altered without their owner’s consent.
The owner of a simulation-experiment description is
able to invite fellow modellers to collaborate on spe-
cific simulation-experiment descriptions. Thus, the
system should make a distinction between anonymous
and authenticated users. Permissions based on roles,
e.g., anonymous visitor, authenticated member, mod-
eller and curator can be used to delegate responsibili-
ties and precisely control access and visibility.
2.1.3 Data Accessibility
Someone using the system should easily be able
to search through the collection of simulation-
experiment descriptions using a variety of search cri-
teria. Finding the the simulation-experiment descrip-
tion he or she wanted, a modeller should be able
to browse through it and, if appropriate, to export
the entire simulation-experiment description to a file
conforming to the SED-ML standard. Over time,
simulation-experiment descriptions should resemble
‘laboratory notebooks’ that contain their entire his-
tories from inception through development to publi-
cation. Additional tools should make it possible to
reproduce automatically simulations based on the ex-
ported SED-ML files.
2.2 Architecture
Based on these system requirements, we implemented
a prototype of the system based on the content man-
agement system Plone
1
(Pastore, 2006). We choose
Plone because its concept of ‘roles’ clarifies responsi-
bilities: this allows modellers to concentrate on creat-
ing their content. Plone’s development tools allowed
us to turn Unified Modeling Language (UML)
2
mod-
els into running Plone code.
Plone is a free and open source content man-
agement system written in Python
3
and built on top
of the Zope
4
application server. It stores all its
information in Zope’s built-in transactional object
database (ZODB) (Fulton, 2000). We have developed
1
www.plone.org
2
www.omg.org/spec/UML/2.1.2
3
www.python.org
4
www.zope.org
our simulation-experiment description repository by
modelling a set of SED-ML content types in UML.
Our models are input to ArchGenXML (Auersperg
et al., 2007) - a code generating tool will turn our
UML models into a Plone add-on product that we in-
stall in a Plone portal running on the Zope server.
2.2.1 Performance
The number of SEDs a repository can hold is limited
only by the physical capacity of the server on which
repository runs.
The Oscillation to Chaos (O2C) SED exam-
ple (K
¨
ohn and Le Nov
`
ere, 2008) is a hierarchy of 18
SED objects in our repository that occupy 30K in the
Zope database. O2C’s exported SED-ML representa-
tion, stripped of all its annotations, is a mere 3.5k.
To evaluate the effect of the Varnish cache we used
ApacheBench
5
to collect median performance data
from both the default Apache/Varnish/Plone route,
and directly from Plone via the zeo-clients. Figure 1
shows how efficacious a caching server becomes as
the number of concurrent requests increase.
Figure 1: Median Total Request Time.
2.2.2 Deployment
The SED repository can be installed on any Zope ap-
plication server running Plone 3. We deploy Plone
on a shared virtual server with two quad-core Xeon
E5405 processors and 4GB of RAM.
Plone is configured in a standard production en-
vironment (Aspeli, 2007) with an Apache web server
acting as a reverse proxy for a Varnish caching server
that balances requests over five zeo clients connected
to a single Zope application server.
Our memory footprint is less than 100MB for the
Zope application server and an average of 300MB for
each zeo-client.
5
httpd.apache.org/docs/2.0/programs/ab.html
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
136
2.3 Object Model
As a formal representation of the simulation ex-
periments, we made use of the Simulation Exper-
iment Description Object Model (SED-OM). The
SED-OM is a formal representation of the MIASE
guidelines expressed using the Unified Modeling
Language (UML). From the object model we derive
the Simulation Experiment Description Meta Lan-
guage (SED-ML) schema that represents the grammar
for SED-ML compliant documents. The SED-ML
XML schema is distributed in XSD
6
format - a W3C
7
consortium standard for defining an XML Document.
Beginning with the SED-OM schema we reverse-
engineered our own object-model - annotated with
Plone supplied tagged-values that specify how data
members look and behave in the Plone UI. For this
purpose, we chose to use an XML editor Named Oxy-
gen
8
and a UML editor named Poseidon
9
. Our repos-
itory and its custom content types are based on the
Level 1 Version 1 (Draft) version of the SED-ML
schema (Bergmann, 2010).
Casting the SED-ML XML schema as a set of
custom content types that represent the objects in the
SED-OM allows nontechnical users to use the content
types as building blocks to create and manage Simu-
lation and Experiment Descriptions. Modelers are not
confronted with complex XML syntax, but create and
manage simulation-experiment descriptions through a
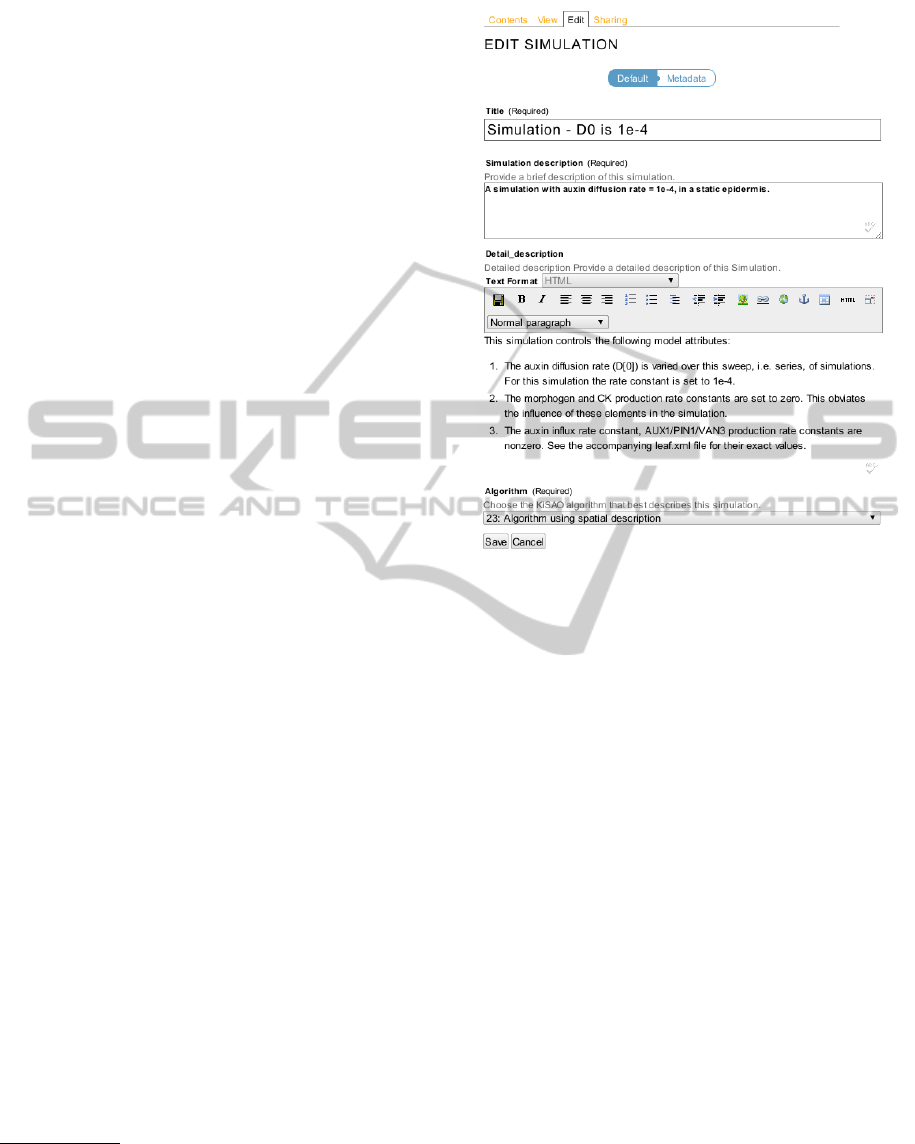
set of clear and concise forms. Figure 2 shows the
Simulation edit form. Its data fields conform to the
SED-ML XML schema simulation element. The de-
tailed description field uses Plone’s own powerful rich
editor with text formatting and image and link inser-
tion abilities. A simulation-experiment description’s
data are stored in such a way that it can be exported
in SED-ML compliant XML.
2.3.1 Code Generation
To generate Plone add-on modules from the UML
class model, we used ArchGenXML
10
. Arch-
GenXML is a code-generator for Plone. Archetypes
is a framework for building content objects based on
schemas and provides features such as automatically
generating editing and presentation views, assigning
unique content Ids, registering content types with
various Plone management tools and storage trans-
parency. Our newly generated content types behave
6
www.w3.org/XML/Schema
7
www.w3c.org
8
www.oxygenxml.com
9
www.gentleware.com
10
plone.org/products/archgenxml
Figure 2: Simulation Edit Form.
like native content types in that they adhere to Plone’s
document workflows and security model.
ArchGenXML automatically generates the edit-
ing and presentation views for each new content type.
In the UML model, we annotate class member data
with tagged-values that describe how it should look
and behave. For example, the standard stipulates that
the top-level simulation-experiment description class
must have an integer version number. The tagged-
values assigned to it indicate that it is a required field,
that it must be an integer, what its label and descrip-
tion text are and its default value.
Tagging simulation-experiment descriptions with
standard and domain meta-information will facilitate
their organisation and classification while simultane-
ously providing rich search criteria. Plone imple-
ments the Dublin Core (Weibel et al., 1998) meta-
data - a small set of text elements through which
Plone content types can be described and catalogued.
Their values are indexed in Plone’s internal cata-
logue and can be used in searches. In addition to
the Dublin Core stipulated ’description’ element, each
simulation-experiment description content type has a
detailed description field in the form of a rich text ed-
itor, that the modeller can use for writing prose de-
scriptions of the simulation experiments.
The simulation-experiment description object
model specific two vocabularies: a model’s encod-
A WEB-BASED REPOSITORY OF REPRODUCIBLE SIMULATION EXPERIMENTS FOR SYSTEMS BIOLOGY
137

ing formalism and a simulation’s algorithm desig-
nation. The former is a simple list including e.g.,
SBML, CellML and C++. The latter describes the
Kinetic Simulation Algorithm Ontology (KiSAO)
11
encoded in the vocabulary description markup lan-
guage (VDEX)
12
. Both vocabularies, while used by
the repository, are managed separately, which allows
the site administrators to modify the contents of the
vocabularies without adversely effecting the existing
models and simulations that use them.
2.3.2 Collaboration Strategies
Plone’s workflows control the progress a modelling
project through a series of states from inception to
publication. Figure 3 shows the states and transi-
tions in a restricted Plone workflow. By customis-
ing these workflows, we can realise a variety of col-
laboration strategies. For example, the current work-
flow for our several simulation-experiment descrip-
tion repositories makes them accessible only to mem-
bers of their respective modelling groups. The work-
flow progressively expands the audience as individual
simulation-experiment descriptions proceed through
a series of approvals, first to other members of the re-
search group, then to portal members, and finally to
the general public.
draf t
start
private
pending
external
internal
show
hide
submit
publish-internally
publish-externally
retract/reject
publish-internally
retract/reject
retract/reject
publish-externally
Figure 3: A Restricted Plone Workflow.
2.3.3 Exporting SED-ML Compliant XML
In order to use the repository for automatically re-
producing simulation experiments, the system must
have facilities to export the simulation-experiment
description to SED-ML compliant XML. We have
11
www.ebi.ac.uk/compneur-srv/kisao
12
www.imsglobal.org/vdex
achieved this by customising the code generated by
ArchGenXML.
The simulation-experiment description class is the
root of a hierarchy that, taken in its entirety, rep-
resents a complete simulation-experiment descrip-
tion. We added custom python code to each class
in this hierarchy to enable their instances to create
an SED-ML representation of themselves and request
their children to do likewise. The code was writ-
ten with Python’s light-weight implementation of the
Document Object Model interface. A document tab
named ’Export this’ appears at the bottom of ev-
ery simulation-experiment description. It is linked
to a browser-view - a special class that links URLs
with specific bits of code in Zope’s component ar-
chitecture. The tab, when clicked, sets a simulation-
experiment description’s export process in motion.
Concluding, we have developed a set of custom
content types that faithfully represent the SED-ML
XML schema, which can be installed into a Plone por-
tal running on a Zope application server customised to
local needs.
2.4 Usage
2.4.1 Data Entry
The user interacts with our system via a Plone-based
web interface. Each SED-ML content type has its
own presentation and editing forms. Creating a
simulation-experiment description consists of navi-
gating to the repository and clicking on the ‘Add
SED’ menu button. A new simulation-experiment de-
scription edit form appears and solicits the required
information, and an appropriate amount of descrip-
tive information. As specified in the schema, the
SED-ML name space and level fields have default val-
ues. Since the simulation-experiment description ob-
ject is the root of a new simulation experiment hierar-
chy, after clicking the simulation-experiment descrip-
tion’s ‘Save’ button, the add items menu presents a
new list of content items the modeller can insert at this
point. This process continues until the simulation-
experiment description is complete.
2.4.2 Searching
Since we specified which information would be in-
dexed while we modelled our SED-ML content types
in UML, searching through the repository is now a
straightforward matter of typing our search criteria
into the search box.
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
138

2.4.3 Reproducing Simulation Experiments
When presented with a complete simulation-
experiment description, especially one written in a
general purpose programming language and not (yet)
reproducible automatically, the modeller can simply
retrieve the recipe from the repository. Section 3 is
a case study describing how to build a simulation-
experiment description whose model is written in
C++.
If a modeller wishes to reproduce the results of a
simulation-experiment description whose models are
written in a dialect of XML, e.g., SBML, and whose
simulations are, e.g., uniform time course, he or she
can export the simulation-experiment description in
SED-ML compliant XML directly by clicking on the
simulation-experiment description’s ‘Export this’ tab.
The repository will return a file whose name is derived
from the simulation-experiment description’s Id. The
modeller can use this file as input to one or another
simulation system such as COPASI (Hoops et al.,
2006).
2.4.4 In Conclusion
The repository is a resource with which a modeller
can fully describe a simulation-experiment descrip-
tion recipe. The SED-ML compliant content types
assist in this by soliciting and validating information
as it is entered. Content and meta-data are indexed
automatically and available as search criteria. A com-
pleted simulation-experiment description should con-
tain the descriptive information to allow the simula-
tion experiment to be repeated manually, and if appro-
priate its contents can be exported in SED-ML com-
pliant XML to be rerun automatically.
3 EXAMPLE SED COMPOSITION
To demonstrate how to compose a SED, we have cho-
sen an experiment that uses the VirtualLeaf (Merks
et al., 2011), an open-source framework for cell-
based modelling of plant tissue growth and develop-
ment. This particular experiment shows how auxins,
a family of plant growth hormones, can accumulate in
growing plant tissue to form bulbous patterns.
The VirtualLeaf distribution contains the Virtual-
Leaf framework software, various dynamical models
and sample simulation data; so acquiring, installing
and running the experiment is simple and comprises
only a few steps.
1. Download the VirtualLeaf Framework source
bundle from its Google Code repository.
2. Compile and install the VirtualLeaf software.
3. Choose a dynamical model to use, i.e. auxin
growth.
4. Choose the simulation data describing the experi-
ments initial conditions.
5. Run the experiment.
6. Aggregate the resulting experiment snapshots into
a single animation.
In a simulation experiment description the informa-
tion in these various steps is distributed over several
SED-ML elements as follows.
• The SED contains two Model elements. The first
model describes the VirtualLeaf framework. The
second model describes the auxin growth model
plug-in that describes the dynamic character of
growing cells. Both models contain unique identi-
fiers naming the particular version of the Virtual-
Leaf software used in the simulation experiment
and its location in a googlecode repository from
where the source code can be downloaded.
• A single Simulation element contains a leaf de-
scription file that describes the initial configura-
tion of cells and their various properties; it also
contains a KiSAO designation of the type of algo-
rithm used.
• A single Task element associates the models with
the simulation as a single experiment.
• A single DataGenerator element describes how
to aggregate the experiment’s results, in this case
screen shots, into an animation; it also includes a
shell script that performs the aggregation.
• A single Output element describes the experi-
ments results, and points to a the dataGenerator
that tell how to reproduce them.
Figure 4 shows the repository’s output element
view of the animation generated by concatenating the
VirtualLeaf’s multi-screenshot output.
A screencast demonstrating how to build these el-
ements is available on our Repository’s public web-
site: sed.project.cwi.nl.
3.1 Run Simulations Automatically
Currently, only XML-based, e.g., SBML, simulations
can be reproduced automatically; using tools like CO-
PASI. For the moment, our SEDs whose models are
written in general-purpose languages contain descrip-
tive information in each SED element to help the
reader understand the simulation and then run it man-
ually.
A WEB-BASED REPOSITORY OF REPRODUCIBLE SIMULATION EXPERIMENTS FOR SYSTEMS BIOLOGY
139

Future versions of the repository will read a SED’s
resulting SED-ML file and generate scripts to repro-
duce the simulation (semi-) automatically using an
approach similar to Python’s buildout (Aspeli, 2007)
system where configuration recipes preform common
tasks such as downloading, compiling and installing
a software source distribution or executing a program
together with its inputs.
4 DISCUSSION
In this paper, we have described a web-based
repository that assists modellers in creating
simulation-experiment descriptions that conform
to the Simulation Experiment Description Markup
Figure 4: Repository Output Element View of a VirtualLeaf
Animation.
Language (SED-ML) - a format for the imple-
mentation of the MIASE guidelines. Modelers
can build simulation-experiment descriptions in an
intuitive way, and annotate them with descriptions,
experimental data and domain meta-information.
This information is indexed automatically and made
available as search criteria. Finally, the contents of
simulation-experiment descriptions can be exported
in SED-ML compliant XML.
Our web-based simulation experiment repository
allows modellers to create and manage simulation-
experiment descriptions in a way analogous to keep-
ing a laboratory notebook. Models can be described
in the context of real simulations at an appropriate
level of abstraction and detail. The entire prove-
nance of a simulation experiment can be preserved
and retraced. Ideally, published simulation experi-
ment will refer to simulation-experiment descriptions
in our repository that are machine-executable and
human-readable recipes that allow modellers to repro-
duce simulations and build upon the published results.
The benefit to the modellers that they have at
their disposal a repository of qualified simulation-
experiment descriptions that describe models in the
context of real simulations. Their detailed technical
specification and associated descriptive information
will help reduce the time and effort needed to repro-
duce simulation results, and provide an excellent start
for reusing and extending models.
Maintaining a proper laboratory notebook re-
quires a great deal of discipline. Adequately anno-
tating a simulation-experiment description is equally
difficult, if not more. For while a laboratory note-
book may be a private work record, it is our inten-
tion that once a simulation study has been published
its simulation-experiment descriptions in our repos-
itory will be read and used by many. The danger
for simulation-experiment descriptions, like any doc-
umentation, is that they will be left incomplete.
While our repository can export SED-ML com-
pliant XML, it cannot (yet) convert it back into a
SED-ML object hierarchy. We are contemplating
writing an import facility to read SED-ML files into
our repository. We would also like to update our
Simulation Experiment Description Object Model to
conform to the SED-ML Level 1 Version 1 (Final).
Currently, only XML-based, e.g., SBML, simulations
can be reproduced automatically; future versions will
also implement scripts that (semi-)automatically re-
produce simulations implemented in general-purpose
languages, e.g., using an approach similar to Python’s
buildout system.
We hope that modellers will be motivated to write
reproducible simulation-experiment descriptions us-
SIMULTECH 2011 - 1st International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
140

ing our repository when they experience the benefits
of having full and immediate on-line access to their
work.
ACKNOWLEDGEMENTS
This work was (co)financed by the Netherlands Con-
sortium for Systems Biology (NCSB) which is part
of the Netherlands Genomics Initiative / Netherlands
Organisation for Scientific Research.
We would also like to thank our colleagues in the
NCSB-NISB Biomodeling and Biosystems Analysis -
Life Sciences Group at CWI for their cooperation in
developing and testing the repository.
REFERENCES
Aspeli, M. (2007). Professional Plone Development. Packt.
Auersperg, P., Klein, J., and van Rees, R. (2007). Arch-
GenXML code generator.
Bergmann, F. (2010). Simulation Experiment Descrip-
tion Markup Language (SED-ML): Level 1 Version
1 (Draft).
Fulton, J. (2000). Introduction to the Zope Object Database.
In Proceedings of the 8th International Python Con-
ference.
Hines, M., Morse, T., Migliore, M., Carnevale, N., and
Shepherd, G. (2004). ModelDB: a database to sup-
port computational neuroscience. Journal of Compu-
tational Neuroscience, 17(1):7–11.
Hoops, S., Sahle, S., Gauges, R., Lee, C., Pahle, J., Simus,
N., Singhal, M., Xu, L., Mendes, P., and Kummer,
U. (2006). COPASI–a COmplex PAthway SImulator.
Bioinformatics, 22(24):3067.
Knuth, D. (1984). Literate programming. The Computer
Journal, 27(2):97.
K
¨
ohn, D. and Le Nov
`
ere, N. (2008). SED-ML–An XML
Format for the Implementation of the MIASE Guide-
lines. In Computational Methods in Systems Biology,
pages 176–190. Springer.
Le Nov
`
ere, N., Bornstein, B., Broicher, A., Courtot, M.,
Donizelli, M., Dharuri, H., Li, L., Sauro, H., Schilstra,
M., Shapiro, B., et al. (2006). BioModels Database: a
free, centralized database of curated, published, quan-
titative kinetic models of biochemical and cellular sys-
tems. Nucleic Acids Research, 34(suppl 1):D689.
Merks, R. M. H., Guravage, M., Inz
´
e, D., and Beemster, G.
T. S. (2011). Virtualleaf: an open source framework
for cell-based modeling of plant tissue growth and de-
velopment. Plant Physiology, 155(656):666.
Pastore, S. (2006). Web content management systems: us-
ing plone open source software to build a website for
research institute needs. In Digital Telecommunica-
tions, , 2006. ICDT ’06. International Conference on,
page 24.
Sivakumaran, S., Hariharaputran, S., Mishra, J., and Bhalla,
U. (2003). The Database of Quantitative Cellular
Signaling: management and analysis of chemical ki-
netic models of signaling networks. Bioinformatics,
19(3):408.
Weibel, S., Kunze, J., Lagoze, C., and Wolf, M. (1998).
Dublin core metadata for resource discovery. Internet
Engineering Task Force RFC, 2413.
Yu, T., Lawson, J. R., and Britten, R. D. (2009). A dis-
tributed revision control system for collaborative de-
velopment of quantitative biological models. In Mag-
jarevic, R., Lim, C. T., and Goh, J. C. H., editors, 13th
International Conference on Biomedical Engineering,
volume 23 of IFMBE Proceedings, pages 1908–1911.
Springer Berlin Heidelberg.
Yu, T., Lloyd, C., Nickerson, D., Cooling, M., Miller, A.,
Garny, A., Terkildsen, J., Lawson, J., Britten, R.,
Hunter, P., et al. (2011). The Physiome Model Repos-
itory 2. Bioinformatics.
A WEB-BASED REPOSITORY OF REPRODUCIBLE SIMULATION EXPERIMENTS FOR SYSTEMS BIOLOGY
141
