
APPLICATION OF GENOME LINGUISTIC APPROACHES FOR
IDENTIFICATION OF GENOMIC ISLAND IN BACTERIAL
GENOMES AND TRACKING DOWN THEIR ORIGINS
Genome Linguistics to Visualize Horizontal Gene Exchange
Oliver Bezuidt, Kingdom Mncube and Oleg N. Reva
University of Pretoria, Department Biochemistry, Bioinformatics and Computational Biology Unit
0002, Pretoria, South Africa
Keywords: Genome linguistics, k-Mer statistics, Oligonucleotide usage pattern, Genomic island, Horizontal gene
exchange, Pathogenicity.
Abstract: With more sequences of complete bacterial genomes getting public availability the approaches of genome
comparison by frequencies of oligonucleotides (k-mers) known also as the genome linguistics are becoming
popular and practical to resolve problems which can not be tackled by the traditional sequence comparison
tools. In this work we present several innovative approaches based on k-mer statistics for detection of
inserts of genomic islands and tracing down the ontological links and origins of mobile genetic elements.
637 bacterial genomes were analyzed by SeqWord Sniffer program that has detected 2,622 putative
genomic islands. These genomic islands were clustered by DNA compositional similarity. A stratigraphic
analysis was introduced that allows distinguishing between new and old genomic inserts. A method of
reconstruction of donor-recipient relations between micro-organisms was proposed. The strain E. coli TY-
2,482 isolated from the latest deadly outbreak of a haemorrhagic infection in Europe in 2011 was used for
the case study. It was shown that this strain appeared on an intersection of two independent fluxes of
horizontal gene exchange, one of which is a conventional for Enterobacteria stream of vectors generated in
marine gamma-Proteobacteria; and the second is a new channel of antibiotic resistance genomic islands
originated from environmental beta-Proteobacteria.
1 INTRODUCTION
Recurrent outbreaks of pathogens armed with new
virulence factors and broad range antibiotic
resistance gene cassettes demonstrate our ignorance
on the principles of horizontal gene exchange and
the evolution of pathogenic bacteria. Ontology and
phylogeny of laterally transferred genetic elements
are difficult to investigate, let alone the predictions
of their insertion sites in hosts chromosomes. DNA
and protein sequence similarity comparison by blast
is generally used to track the origins of genomic
islands (GIs) but this approach has many limitations.
Horizontally transferred genes are highly mutable
and similarities in their DNA sequences quickly
disappear. Protein sequence similarity reflects
mostly functional conservations rather than the
phylogenetic relations between GIs. Moreover, the
majority of genes found in GIs are hypothetical and
in many instances falsely predicted. Multiple genes
which are of great importance for mobility of
plasmids and phages quickly become a wreck after
integration into chromosomes due to multiple
mutations and fragmentations.
Bacterial species are variable in their overall GC
content but the genes in genomes of particular
species are fairly uniform with respect to their base
composition patterns and frequencies of
oligonucleotides. DNA compositional comparison of
bacterial genomes showed that horizontally acquired
genes display features that are distinct from those of
their recipient genomes (Van Passel, 2006). Based
on these observations we developed and utilized
several innovative genome linguistics approaches to
improve GI detection in bacterial genomes. They are
practical also for reconstraction of the ontological
links and donor-resipient relations between
microorganisms and their mobilomes.
118
Bezuidt O., Mncube K. and N. Reva O..
APPLICATION OF GENOME LINGUISTIC APPROACHES FOR IDENTIFICATION OF GENOMIC ISLAND IN BACTERIAL GENOMES AND TRACKING
DOWN THEIR ORIGINS - Genome Linguistics to Visualize Horizontal Gene Exchange.
DOI: 10.5220/0003704201180123
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2012), pages 118-123
ISBN: 978-989-8425-90-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

2 RESULTS
2.1 Oligonucleotide Usage Pattern
Concept
Statistical parameters of frequencies of k-mers in
natural DNA sequences and a definition of the
oligonucleotide usage (OU) pattern were given in
our previous publications (Reva, 2005); (Ganesan,
2008). OU pattern was denoted as a matrix of
deviations
[
1…
N]
of observed from expected counts
for all possible permutations of oligonucleotides
(words) of length N. In this work tetranucleotides
were used, thus N = 256. Words are distributed in
sequences logarithmically and the deviations of their
frequencies from expectations may be found as
follows:
1ln
ln
6
22
222
222
...1
|...10|...1
0|...1|...1|...1
0|...1|...1|...1
eNN
NobsNeN
NeNobsN
CC
CCC
CCC
N
w
(1)
where
n
is any nucleotide A, T, G or C in the N-
long word; C
[
1…
N]|obs
is the observed count of a
word [
1
…
N
]; C
[
1…
N]|e
is its expected count and
C
[
1…
N]|0
is a standard count estimated from the
assumption of an equal distribution of words in the
sequence: C
[
1…
N]|0
= L
seq
4
-N
. In this work
C
[
1…
N]|e
frequencies were calculated by using 0-
order Markov model, i.e. by normalization to the
frequencies of nucleotides.
The distance D between two patterns was
calculated as the sum of absolute subtractions of
ranks of identical words after ordering of the words
by
[
1…
N]
values (equation 1) in patterns i and j as
follows:
minmax
min
4
,,
100(%)
DD
Drankrank
D
N
w
jwiw
(2)
Application of ranks instead of relative
oligonucleotide frequencies made the comparison of
OU patterns less biased to the sequence length
provided that the sequences are longer 5 kbp for a
reliable statistics of tetranucleotide frequencies
(Reva, 2005).
Pattern skew (PS) is a particular case of D where
patterns i and j are calculated for the same DNA but
for direct and reversed strands, respectively.
D
max
= 4
N
(4
N
– 1)/2 and D
min
= 0 when calculating
a D, or, in a case of PS calculation, D
min
= 4
N
if N is
an odd number, or D
min
= 4
N
– 2
N
if N is an even
number due to the presence of palindromic words.
Relative variance of an OU pattern (RV) was
calculated by the following equation:
0
4
2
14
N
w
w
N
RV
(3)
where N is the word length; Δ
2
w
is the square of a
word w count deviation (see equation 1); and σ
0
is
the expected standard deviation of the word
distribution in a randomly generated sequence which
depends on the sequence length (L
seq
) and the word
length (N):
seq
N
L
4
02.0
0
(4)
GRV is a particular case of RV when expected counts
of words are calculated based on the frequencies of
nucleotides estimated for the complete genome.
2.2 Identification and Clustering of
Genomic Islands
Each genome may be characterized by a unique
pattern of frequencies of oligonucleotides (Abe,
2003); (Reva, 2005;). Foreign DNA inserts retain
OU patterns of the genomes of origin. Comparison
of OU patterns of genomic fragments against the
entire genome OU pattern reveals areas with
alternative DNA compositions. An algorithm for the
identification of horizontally transferred genomic
elements by superimposition of OU statistical
parameters has been introduced in our previous
publications (Reva, 2005); (Ganesan, 2008). This
algorithm calculates and superimposes the four OU
pattern parameters discussed above: D – distance
between local and global OU patterns; RV and GRV
variances; and PS. Horizontally transferred GIs are
characterized by a significant pattern deviation
(large D), significant increase in GRV associated
with decreased RV and a moderately increased PS
(Ganesan, 2008). Exploitation of all these parameter
allows the discrimination of the putative mobile
genomic elements from other genomic loci with
alternative DNA compositions, namely: multiple
tandem repeats, clusters of genes for ribosomal
proteins and ribosomal RNA, giant genes, etc (Reva,
2005). To facilitate a large scale analysis of bacterial
genomes, a Python utility SeqWord Sniffer was
developed. It is available for download from
www.bi.up.ac.za/SeqWord/sniffer/.
Sniffer was compared to other available tools of
GI identification: IslandPick, SIGI-HMM and
IslandPath (Langille, 2009). The rate of false
negative predictions was determined by testing the
capacities of different programs to predict known
APPLICATION OF GENOME LINGUISTIC APPROACHES FOR IDENTIFICATION OF GENOMIC ISLAND IN
BACTERIAL GENOMES AND TRACKING DOWN THEIR ORIGINS - Genome Linguistics to Visualize Horizontal
Gene Exchange
119

patogenicity GIs from PAI DB (http://www.gem.
re.kr/paidb/about_paidb.php). The rate of false
positive predictions was estimated by counting the
numbers of GIs predicted by one program which
were not confirmed by other programs. To optimize
the Sniffer’s program run settings the factorial
experiment was used. Results of program
comparison are shown in Table 1.
Table 1: Comparison of different programs for GI
identification.
Program False negative rate False positive rate*
Sniffer 0.12 0.44
IslanPick 0.94 0.40
SIGI-HMM 0.41 0.28
IslandPath 0.69 0.43
*It has to be taken into consideration that not all unconfirmed GIs
are false positives. It is assumed that the false positive rates in this
column are at least twice as much as they are.
Sniffer and SIGI-HMM predicted more than 60%
of know pathogenicity GIs, and Sniffer showed the
smallest rate of false positives.
In this work we searched for GIs in a set of 637
bacterial genomes representing different taxonomic
classes. In a total, 2,622 GIs were predicted (visit
http://anjie.bi.up.ac.za/geidb/geidb-home.php for
more information). Then these GIs were clustered
basing on OU pattern similarity. The assumption is
that the GIs which originated from the same source
share compositional similarity. Compositional
similarity was measured as 100% – D. GIs with a
pattern similarity above 75% were often found to
share homologous blocks of DNA sequences. Hence
a pattern similarity index of 75% was chosen as a
threshold for clustering of GIs. In total 1,305
clusters were obtained; however, 1,158 of the total
clusters were singletons. To visualize the relations
between GIs in clusters, an in-house Python script
with Graphviz incorporation and a graph pruning
criterion implementation was used. The pruning
method was implemented as follows: if three nodes
in a graph are interlinked, the edge representing the
smallest similarity percentage gets pruned. The
script consequently determines sequence similarities
between linked GIs by bl2seq algorithm and
generates a graphical output (Fig. 1.)
2.3 Stratigraphic Analysis of Genomic
Islands
Inserts of foreign DNA undergo a process of
amelioration, which influences their OU patterns to
start to reflect the OU pattern of their host
chromosomes overtime (Lawrence, 1997). We
hypothesized that the comparison of GIs of the same
origin which are distributed in organisms that share
similar genomic OU patterns would result in
different D-values relative to the time of their
acquisition. In Fig. 2 the differences in D-values are
depicted by grey colour gradients. The darker colour
gradients depict GIs which have been acquired
recently, while the lighter colours depict ancient
acquisitions.
3 DISCUSSION
DNA molecules encoding functional enzymes,
transcriptional regulators and virulence determinants
are fluxing through the bacterial taxonomic walls.
They endow environmental and clinical strains of
bacteria with new unexpected properties. Lateral
genetic exchange, particularly of drug tolerance
genes has been recognized for a long time; however
the phenomenon of the horizontal gene transfer is
generally obscure. Linguistic approaches based on
the analysis of biased distribution of
tetranucleotides, which were applied in this study,
showed to be instrumental for the identification of
GIs in bacterial genomes; grouping of inserts
originated from one or multiple sources; and also for
the estimation of the approximate time when transfer
events have occurred. This information is relevant to
understanding of the role that the horizontal gene
exchange plays in evolution of new pathogens. The
analysis of the complete genome sequence of the
strain E. coli TY-2482 isolated from the latest
outbreak of entero-aggregative-haemorrhagic
(EAHEC) infection in Europe in 2011 showed that
its extraordinary virulence and lethality are
associated with the virulence determinants located in
horizontally transferred GIs (Brzuszkiewicz, 2011);
(Manrique, 2011). Enterobacteria (Escherichia,
Shigella and Salmonella) share multiple GIs shown
in Fig. 1 and 2 in cells A2-B4, which comprise all
well known enterobacterial pathogenicity GIs (see
PAIDB at www.gem.re.kr/paidb/about_paidb.php).
It was found that the inserts of antibiotic
resistance genes in E. coli TY-2482 and several
Salmonella ontologically are not linked to the major
cluster of the enterobacterial GIs but fall into a
rather versatile group of mobile genetic elements
distributed among beta-Proteobacteria, alpha-
Proteobacteria, Actinobacteria (predominantly
Mycobacteria) and Acidobacteria, which is shown in
Fig. 1 and 2 in cells E1-F2. Many of these GIs are
old inserts but those in gamma-Proteobacteria are
very recent acquisitions.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
120
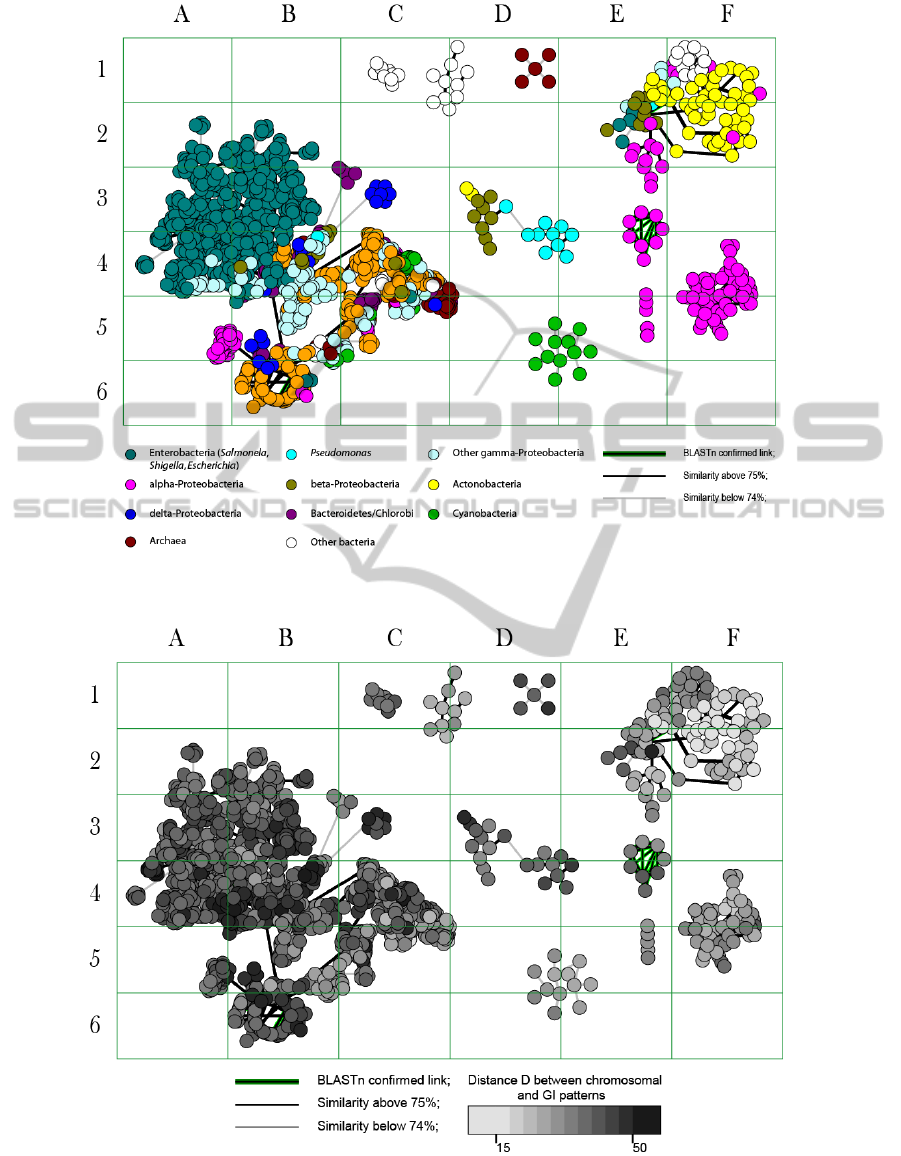
Figure 1: Clusters of GIs from different bacterial classes. Each node represents one GI. For more details visit the interactive
map at http://www.bi.up.ac.za/SeqWord/maps/map.html (tested on Mozilla Firefox 5.0).
Figure 2: Stratigraphic analysis of GI inserts. Each node represents one GI. For more details visit the interactive map at
http://www.bi.up.ac.za/SeqWord/maps/map.html (tested on Mozilla Firefox 5.0).
APPLICATION OF GENOME LINGUISTIC APPROACHES FOR IDENTIFICATION OF GENOMIC ISLAND IN
BACTERIAL GENOMES AND TRACKING DOWN THEIR ORIGINS - Genome Linguistics to Visualize Horizontal
Gene Exchange
121

They show DNA compositional similarity to GIs
from beta-Proteobacteria (Fig. 2). Relatedness
between these GIs was confirmed by the sequence
similarity revealed by blast. Particularly, many of
these GIs contain a large mercury resistance operon,
which might be adopted in Enetrobacteria to
withstand antibiotics. To identify which organisms
may be donors of GIs and which are likely to be
recipients, we developed an algorithm of cross-
comparison of OU patterns of GIs and their hosts.
The result of this analysis for a pair of genomes
Acidovorax ebreus TPSY and S. enterica plasmid
CT18 containing similar GIs with mercury
resistance genes is visualized in Fig. 3.
Fig. 3 shows that these GIs have originated from
Acidovorax lineage and then were donated to
Salmonella. This is in consistence with the fact that
the inserts in Salmonella and Escherichia genomes
are much more recent than those in beta-
Proteobacteria (Fig. 2, cell E2).
Figure 3: Donor recipient relations determined for GIs of
A. ebreus and S. enterica. X shows the distance between
host chromosomes depicted by dark green circles. GIs of
the organisms on the left and right are shown as red and
blue circles, respectively.
Donor-recipient analysis was applied to analyse
the directions of distribution of GIs of the biggest
cluster in cells A2-C6 (Fig. 1). The reconstructed
pathways are shown in Fig. 4. This cluster of GIs
represents the most active mainstream of the
horizontal gene exchange that penetrated the borders
of multiple Archaeal and Eubactarial genera. The
oldest inserts where found in Pyrococcus,
Methanosarcina, Methanospirillum and some other
Archaea, as well as in Enterococcus, Listeria and
Anabaena. Shewanella and Chlorobium played an
important role in a recent transmission of these GIs
towards other bacterial genera. For instance, all
enterobacterial GIs in the cells A2-B4, as well as
GIs of Lactobacillus in cells C4 (Fig. 1) have
originated from the Shewanella lineage. GIs of
Bacillus, Geobacillus, Pelobacter and Geobacter
show compositional similarity to more ancient GIs
of Chrolobium.
The stratigraphic analysis showed that it was not
a single flux of GIs, but a recursive process of
generation of active vectors which then were
transferred along the chain of donor and recipient
organisms. Time series of inserts of similar GIs but
acquired in different time frames may be observed in
different taxonomic groups of this cluster of GIs
(Fig 1 and 2, cells A2-C6). Oscillations of horizontal
gene exchange activity which may result from a
counterbalance between the acquired resistance of
bacteria towards existing mobile vectors and the
generation of new vectors in the environmental
microflora explain recurrent appearance of new
pathogens.
Figure 4: Putative pathways of distribution of GIs through
bacterial genera.
Other clusters of GIs in cells C1, D1-F6 are in
general older inserts of mobile genetic elements.
However, the dormant GIs may be re-activated as it
has happened with the newly acquired GIs of
Salmonella and Escherichia, which we discussed
above (the cell E2 in Fig. 1 and 2). An unusual rise
in mercury resistance marine bacteria was reported
in coastal environment in India from 1997 to 2003
(Ramaiah, 2003). It was concluded that this sharp
rise in mercury tolerance could be linked to the
general ocean pollution by human industrial activity.
A few years later new GIs with the mercury
resistance operons were found in Salmonella and
they were associated with an increased virulence and
antibiotic resistance of the pathogens (Levings,
2007). To conclude, the global ocean pollution has
activated dormant GIs in the environmental micro-
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
122

flora which reached pathogenic enterobacteria. An
interference of GIs from this new channel (the cell
E2) with the conventional oscillations of
pathogenicity GIs of Enterobacteria (the cells A2-C6
in Fig. 1 and 2) led to appearance of the new deadly
E. coli EAHEC in Europe in 2011.
4 CONCLUSIONS
Repeated outbreaks of new pathogens revealed our
ignorance on the principles of horizontal gene
exchange and the evolution of pathogenic bacteria.
The strain E. coli TY-2482 from the latest outbreak
has been quickly isolated and sequenced that
allowed the discovery of its closest relatives but it
failed to resolve the origin of this strain and the way
of its evolution that made impossible for us to
predict upcoming outbreaks in future. Inability to
answer these questions showed limitations of the
current methods of comparative and evolutionary
genomics.
In this work several innovative approaches of
genome linguistics based on the analysis of the
biased distribution of tetranucleotide were
introduced. These methods were used for clustering
of GIs generated from the same source, estimation of
the relative time of GI insertions and reconstruction
of donor-recipient relations. The genome linguistic
approaches gain more credibility when used in
parallel with the traditional methods of sequence
similarity comparison.
It was found that the recurrent appearance of new
pathogens may be associated with the regular
oscillations of GI vectors. Pathogens may gain an
increased virulence when they are reached by GIs
from unusual sources. There is a pressing need to
create a system that will allow the monitoring of
distributions of horizontally transferred GIs, to aid
us in being informed and prepared regarding the
emergence of new pathogens.
ACKNOWLEDGEMENTS
This work was funded by the National Research
Foundation (South Africa) grant #71261 for
National Bioinformatics and Functional Genomics
Programme.
REFERENCES
Abe, T., Kanaya, S., Kinouchi, M., Ichiba, Y., Kozuki, T.,
et al., 2003. Informatics for unveiling hidden genome
signatures. Genome Res. 13: 693-702.
Brzuszkiewicz, E., Thürmer, A., Schuldes, J., Leimbach,
A., Liesegang, H., et al., 2011. Genome sequence
analyses of two isolates from the recent Escherichia
coli outbreak in Germany reveal the emergence of a
new pathotype: Entero-Aggregative-Haemorrhagic
Escherichia coli (EAHEC). Arch. Microbiol.,
doi:10.1007/s00203-011-0725-6.
Ganesan, H., Rakitianskaia, A. S., Davenport, C. F.,
Tümmler, B., Reva, O. N. 2008. The SeqWord
Genome Browser: an online tool for the identification
and visualization of atypical regions of bacterial
genomes through oligonucleotide usage. BMC
Bioinformatics 9: 333.
Langille, M. G., Brinkman, F. S., 2009. IslandViewer: an
integrated interface for computational identification
and visualization of genomic islands. Bioinformatics
25: 664-665.
Lawrence, J. G., Ochman, H., 1997. Amelioration of
bacterial genomes: rates of change and exchange. J.
Mol. Evol. 44: 383-397.
Levings, R. S., Partridge, S. R., Djordjevic, S. P., Hall, R.
M., 2007. SGI1-K, a variant of the SGI1 genomic
island carrying a mercury resistance region, in
Salmonella enterica serovar Kentucky. Antimicrob.
Agents Chemother. 51: 317-323.
Manrique, M., Pareja-Tobes, P., Pareja-Tobes, E., Pareja,
E., Tobes, R. 2011. Escherichia coli EHEC Germany
outbreak preliminary functional annotation using BG7
system. Nut. Preceedings, doi:10.1038/npre.2011.
6001.1.
Ramaiah, N., De, J., 2003. Unusual rise in mercury-
resistant bacteria in coastal environs. Microb. Ecol. 45:
444-454.
Reva, O. N., Tümmler, B., 2005. Differentiation of regions
with atypical oligonucleotide composition in bacterial
genomes. BMC Bioinformatics 6: 251.
Van Passel, M. W., Bart, A., Luyf, A. C., van Kampen, A.
H. van der Ende, A., 2006. The reach of the genome
signature in prokaryotes. BMC Evol. Biol. 6: 84.
APPLICATION OF GENOME LINGUISTIC APPROACHES FOR IDENTIFICATION OF GENOMIC ISLAND IN
BACTERIAL GENOMES AND TRACKING DOWN THEIR ORIGINS - Genome Linguistics to Visualize Horizontal
Gene Exchange
123
