
RULE EXTRACTION FROM MEDICAL DATA WITHOUT
DISCRETIZATION OF NUMERICAL ATTRIBUTES
Juan L. Domínguez-Olmedo, Jacinto Mata, Victoria Pachón and Manuel J. Maña
Escuela Técnica Superior de Ingeniería, Universidad de Huelva, Ctra. Palos de la Frontera SN, Huelva, Spain
Keywords: Data mining, Association rules, Atherosclerosis data.
Abstract: Association rule mining is a popular technique used to find associations between attributes in a dataset.
When using deterministic algorithms, if the attributes have numerical values the usual approach is to
discretize them defining proper intervals. But the discretization can notably affect the quality of the rules
generated. This work presents a method based on a deterministic exploration of the interval search space
without a previous discretization of the numerical attributes. It has been applied to medical data from an
atherosclerosis study. The quality of the obtained rules seems to support this method as a valid alternative
for this kind of rule extraction.
1 INTRODUCTION
In computer science, the field of Knowledge
Discovery in Databases (KDD) treats the problem of
finding useful knowledge from data. It is based on
several stages that try to derive information of
interest from raw data: selection, preprocessing,
transformation, data mining and evaluation (Fayyad,
1996).
One kind of task of application in the data
mining step is association rule learning. This
technique is used to discover associations between
several variables (attributes) in a dataset, which
could be of interest because they might show
unexpected or unknown relationships between the
variables they associate.
Association rules are not predictive but
descriptive. Descriptive mining tasks characterize
the general properties of the data (Han, 2006).
In this work, a deterministic method to generate
association rules without the discretization of
numerical variables is applied to medical data from
an atherosclerosis study.
Concretely, the STULONG dataset has been
used to extract association rules from it. This dataset
holds data from a longitudinal study of the factors of
the atherosclerosis in the population of 1417 middle
aged men (Boudík, 2004).
Atherosclerosis (also known as arteriosclerotic
vascular disease) is a condition in which an artery
wall thickens as a result of the accumulation of fatty
materials such as cholesterol. Among the first
symptom of atherosclerotic cardiovascular disease is
heart attack or sudden cardiac death.
The organization of this paper is as follows. The
next section provides a preliminary on association
rules and some quality measures. Section 3 describes
the method employed. In section 4 the experimental
results are shown. Finally, section 5 provides some
conclusions.
2 ASSOCIATION RULES
Association rule learning is a popular technique used
to find associations between several attributes in a
dataset. An association rule takes the form A C,
where A and C express conditions on attributes of
the dataset, respectively, the antecedent and the
consequent of the rule.
At the beginning of the use of association rules
for data mining tasks, its application was mainly to
transactional format data, as for market basket
datasets (Agrawal, 1993), where the aim was to
discover regularities between products in large scale
transaction data from supermarkets, as the basis for
decisions about marketing activities.
In addition to this early application, association
rules are employed today in many application areas
including scientific data analysis, Web usage mining
or bioinformatics.
397
L. Domínguez-Olmedo J., Mata J., Pachón V. and J. Maña M..
RULE EXTRACTION FROM MEDICAL DATA WITHOUT DISCRETIZATION OF NUMERICAL ATTRIBUTES.
DOI: 10.5220/0003784603970400
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2012), pages 397-400
ISBN: 978-989-8425-88-1
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

Rules potentially of interest (strong rules) are
those ones with “good” measures of their support
and confidence. The support evaluates the number of
instances (also it can be shown as a percentage) in
which both the antecedent and the consequent of the
rule hold. The confidence is the quotient between the
support of the rule and the number of instances in
which the antecedent holds (the accuracy of the
rule):
conf (A C) = supp (A C) / supp (A)
(1)
So, the extraction of association rules is based on the
search for those ones satisfying minsup and minconf,
thresholds for the minimum support and minimum
confidence of a rule to be considered interesting.
Another measure of the interestingness of a rule is
the lift (Brin, 1997), which measures how many
times more often the antecedent and consequent hold
together in the dataset than would be expected if
they were statistically independent. It can be
computed by the quotient between the confidence of
the rule and the probability of the consequent:
lift (A
C) = conf (A
C) / P (C)
(2)
Several methods have been developed to treat this
problem, mainly variations of the Apriori algorithm
(Bodon, 2005); (Borgelt, 2003).
Nevertheless, applying these methods to data
containing numerical (quantitative) attributes, the
common case in many datasets, can not be done
directly (Srikant, 1996).
The typical approach to the treatment of
numerical attributes is by a previous discretization of
them. But when discretization is applied to
numerical attributes in association rule mining, it is
not possible to employ the usual methods of
application in classification models, such as those
based on the information theory (Tsai, 2008); (Lee,
2007). Instead, unsupervised discretization methods
such as equi-width or equi-frequency have to be used
(Liu, 2002).
3 METHOD EMPLOYED
In this work, we have used a method based on a
deterministic approach to treat the problem of
generating association rules without a previous
discretization of the numerical attributes.
In contrast to the typical deterministic fashion of
obtaining quantitative association rules, that is, by
previously discretizing those attributes, the method
employed is based on a dynamic generation of
intervals for each numerical attribute, searching for
valid rules satisfying the thresholds minsup and
minconf.
The method also employs auxiliary data
structures and certain optimizations to reduce the
search and improve the quality of the rules extracted.
Following are described its main features:
The bounds of the intervals of the numerical
attributes are restricted to existing values in the
dataset.
To have an efficient way of generating the
interval bounds and calculating the rule quality
measures, several auxiliary tables are used. The
bounds are going to be searched in the range [1, n],
where n is the number of instances of the dataset.
And at the end, the bounds are transformed into the
original values for each attribute.
To reduce the number of rules generated,
although probably discarding some rules of good
quality, a parameter delta [0, 1] is also used to
control the exhaustivity of the rule searching
process.
4 APPLICATION TO MEDICAL
DATA
The method described has been applied to medical
data from an atherosclerosis study.
STULONG is a dataset concerning the twenty
years lasting longitudinal study of the factors of the
atherosclerosis in the population of 1417 middle
aged men. Its table Entry holds results of the entry
examinations of each patient, and the table Death
holds data concerning the death of 389 patients. In
the experiments the table Entry_Dead has been used.
This table is the join of Entry and Death tables, with
some previous preprocessing (Salleb, 2004).
First, we have searched for rules associating
attributes of the group “Physical examination” (BMI
“Body Mass Index”, SYST “Blood pressure
systolic”, DIAST “Blood pressure diastolic”, TRIC
“Skin fold triceps” and SUBSC “Skin fold
subscapularis”) with attributes of the group
“Biochemical examination” (CHLST “Cholesterol”,
TRIGL “Triglycerides”, MOC_SUC “Urine sugar”
and MOC_ALB “Urine albumen”).
We have run the algorithm searching for those
kinds of rules, with the parameters minsup = 29
(2%), minconf = 0.5 and delta = 0.2. A selection of
the rules generated is shown in Table 1, based on
rules with high values of lift or with both high
confidence and not-low lift.
HEALTHINF 2012 - International Conference on Health Informatics
398
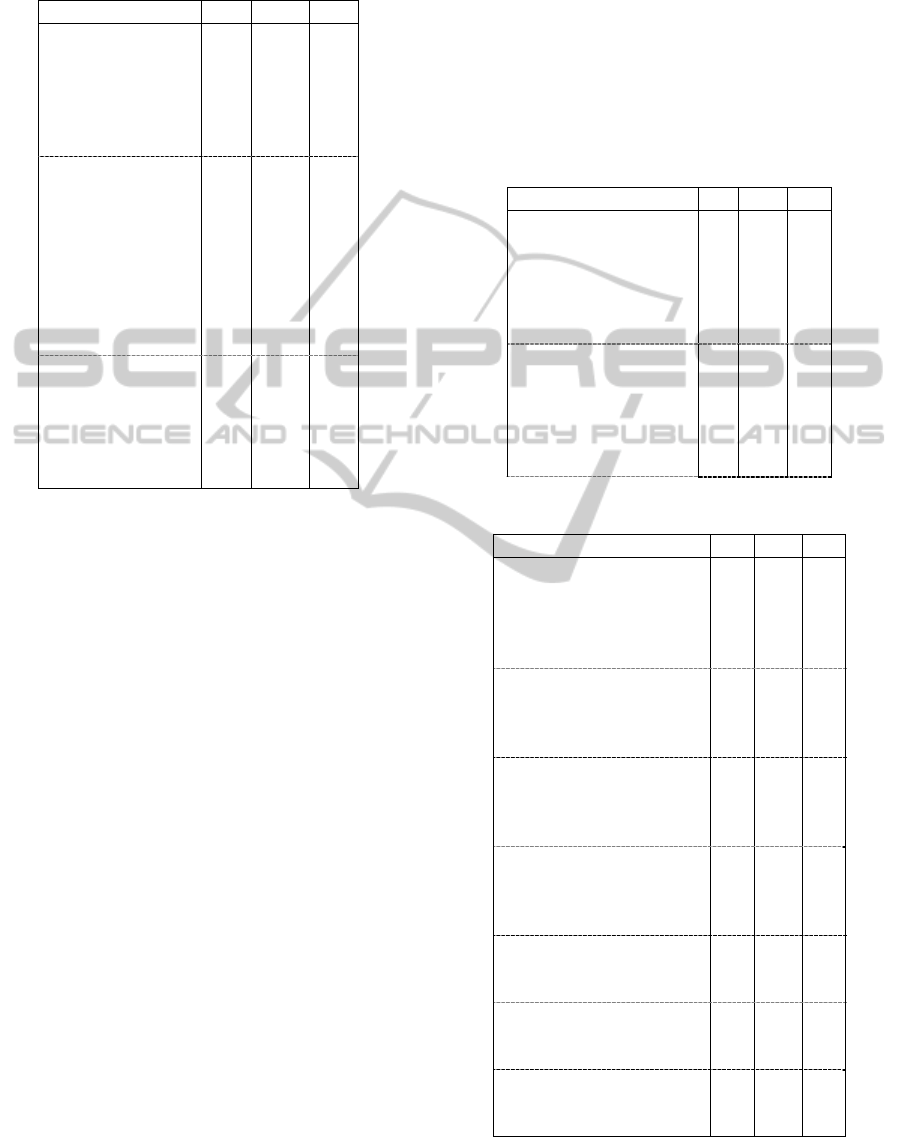
Table 1: A selection of rules associating the groups of
attributes “Physical examination” and “Biochemical
examination”.
Rule Sup Conf Lift
CHLST [112, 242]
TRIGL [28, 71]
BMI [19.05, 28.41]
TRIC [3, 35]
SUBSC [7, 16]
31 0.79 2.3
DIAST [82, 125]
BMI [24.11, 27.36]
TRIC [11, 35]
SUBSC [16, 70]
MOC_SUC = ‘no’
MOC_ALB = ‘no’
CHLST [221, 250]
TRIGL [105, 274]
29 0.51 2.6
TRIC [7, 12]
SUBSC [32, 49]
MOC_SUC = ‘no’
CHLST [211, 300]
TRIGL [103, 350]
29 1.00 2.0
The first of the shown rules states that “79% of
patients having a cholesterol measure in the interval
[112, 242] and a triglycerides measure in the
interval [28, 71], also had a value of BMI in the
interval [19.05, 28.41], a value in the interval [3,
35] for skin fold triceps and a value in the interval
[7, 16] for skin fold subscapularis. The conditions of
the consequent occur 2.3 more times in the group of
patients holding the conditions of the antecedent
than in the whole group of studied patients”.
The last shown rule states that “all patients with
a value in the interval [7, 12] for skin fold triceps
and a value in the interval [32, 49] for skin fold
subscapularis, had also no urine sugar, a
cholesterol measure in the interval [211, 300] and a
triglycerides measure in the interval [103, 350].
The conditions of the consequent occur 2 more times
in the group of patients holding the conditions of the
antecedent than in the whole group of studied
patients”.
We have also searched for rules associating the
attribute
DEATH? with the attributes ALCO_CONS
(“Alcohol consumption”), TOBA_CONS (“Tobacco
consumption”), TOBA_DURA (“Smoking
duration”), MOC_SUC (“Urine sugar”), MOC_ALB
(“Urine albumen”), CHLST (“Cholesterol”), TRIGL
(“Triglycerides”), SYST (“Blood pressure systolic”),
DIAST (“Blood pressure diastolic”) and BMI (“Body
Mass Index”).
The attribute ALCO_CONS measures the volume
of alcohol ingested, taking into account three
factors: the equivalent amount of alcohol, the type of
alcohol, and the patient’s weight (as a normalizing
factor).
Table 2 presents a selection of the rules obtained
after searching with the parameters minsup = 29
(2%), minconf = 0.5 and delta = 0.2.
Table 2: A selection of rules associating the attribute
DEATH?
Antecedent Sup Conf Lift
ALCO_CONS [1, 1.2]
TOBA_DURA = 20
CHLST [197, 273]
SYST [149, 220]
DEATH? = yes
30 0.73 2.7
TOBA_CONSO = 0
CHLST [190, 261]
SYST [ 80, 133]
BMI [22.64, 27.41]
DEATH? = no
52 1.00 1.4
Table 3: Analysis of a rule regarding patient’s death.
Antecedent Sup Conf Lift
ALCO_CONS [1.12, 1.69]
TOBA_CONSO = 1.25
SYST [140, 220]
DEATH? = yes
30 0.65 2.4
ALCO_CONS [1.12, 1.69]
TOBA_CONSO = 1.25
DEATH? = yes
81 0.43 1.6
ALCO_CONS [1.12, 1.69]
SYST [140, 220]
DEATH? = yes
71 0.41 1.5
TOBA_CONSO = 1.25
SYST [140, 220]
DEATH? = yes
47 0.56 2.0
ALCO_CONS [1.12, 1.69]
DEATH? = yes
214 0.27 1.0
TOBA_CONSO = 1.25
DEATH? = yes
132 0.38 1.4
SYST [140, 220]
DEATH? = yes
127 0.39 1.4
RULE EXTRACTION FROM MEDICAL DATA WITHOUT DISCRETIZATION OF NUMERICAL ATTRIBUTES
399

Finally, a kind of analysis about the “additive”
effect of conditions in patient’s death has been done.
For that, the first of the rules shown in Table 3 has
been chosen from the rules generated previously. It
associates alcohol consumption, tobacco
consumption and systolic blood pressure with
patient’s death. This rule expresses that “65% of the
patients with an alcohol consumption in [1.12,
1.69], smoking more than 20 cigarettes/day and with
a systolic blood pressure in [140, 220], were dead”.
To compare the effect of those conditions, alone
and in pairs, rules having the desired conditions have
been selected, and their quality measures are shown
in Table 3.
An analysis of the rules indicates that although
the condition associated to alcohol consumption is
less correlated to death (with a lift value of 1) than
the other two conditions evaluated, when added to
the combination of tobacco consumption and blood
pressure, it increases the confidence from 0.56 to
0.65.
5 CONCLUSIONS
In this work, medical data from an atherosclerosis
study has been used to extract association rules from
it.
Association rules can express unknown
knowledge present in data, in the form of
relationships between the values of the variables.
The method employed is based on a
deterministic approach that generates association
rules without a previous discretization of the
numerical attributes. Discretization can notably
affect the quality of the rules generated, and it is
usually difficult to know the best discretization
technique to apply it to a deterministic algorithm for
a particular dataset.
A variety of rules has been obtained, with good
values of their quality measures, what seems to
support the method employed as a valid way to
generate association rules without a previous
discretization of the numerical attributes.
Also, a particular analysis of a selected rule has
been performed. The rule associates some conditions
with the death of patients object of the study.
ACKNOWLEDGEMENTS
This work was partially funded by the Spanish
Ministry of Science and Innovation, the Spanish
Government Plan E and the European Union through
ERDF (TIN2009-14057-C03-03).
REFERENCES
Agrawal, R., Imielinski, T., Swami, A., 1993. Mining
Association Rules between Sets of Items in Large
Databases. In ACM SIGMOD ICMD, pp. 207-216.
ACM Press.
Bodon, F., 2005. A Trie-based APRIORI Implementation
for Mining Frequent Item Sequences. In 1st
International Workshop on Open Source Data Mining:
Frequent Pattern Mining Implementations, Chicago,
Illinois, pp. 56–65. ACM Press.
Borgelt, C., 2003. Efficient Implementations of Apriori
and Eclat. In Workshop on Frequent Itemset Mining
Implementations. CEUR Workshop Proc. 90, Florida.
Boudík, F., Tomečková, M., Bultas, J., 2004. STULONG
medical project. http://euromise.vse.cz/challenge2004.
Prague.
Brin, S., Motwani, R., Ullman, J.D., Tsur, S., 1997.
Dynamic Itemset Counting and Implication Rules for
Market Basket Data. In Proc. of the ACM SIGMOD
1997, pp. 265-276.
Fayyad, U., Piatetsky-Shapiro, G., Smyth, P., 1996. From
Data Mining to Knowledge Discovery in Databases.
AI Magazine, Vol. 17, pp. 37-54.
Han, J., Kamber, M., 2006. Data Mining: Concepts and
Techniques. Morgan Kaufmann, San Francisco.
Lee, C.-H., 2007. A Hellinger-based Discretization
Method for Numeric Attributes in Classification
Learning. Knowledge-Based Systems, 20(4), 419-425.
Liu, H., Hussain, F., Tan, C., Dash, M., 2002.
Discretization: An Enabling Technique. Data Mining
and Knowledge Discovery, 6(4), 393-423.
Salleb, A., Turmeaux, T., Vrain, C., Nortet, C., 2004.
Mining Quantitative Association Rules in a
Atherosclerosis Dataset. Contribution to the PKDD
Discovery Challenge 2004, http://www.univ-
orleans.fr/lifo/Members/salleb/Challenge2004.
Srikant, R., Agrawal, R., 1996. Mining Quantitative
Association Rules in Large Relational Tables. In Proc.
of the ACM SIGMOD 1996, pp. 1-12.
Tsai, C.-J., Lee, C.-I., Yang, W.-P., 2008. A Discretization
Algorithm Based on Class-Attribute Contingency
Coefficient. Information Science, 178(3), 714-731.
HEALTHINF 2012 - International Conference on Health Informatics
400
