
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN
NEUROIMAGING
Signed Sensitivity Maps
Peter M. Rasmussen
1,2
, Tanya Schmah
3
, Kristoffer H. Madsen
1,4
, Torben E. Lund
2
,
Grigori Yourganov
5,6
, Stephen C. Strother
5,7
and Lars K. Hansen
1
1
DTU Informatics, Technical University of Denmark, Kgs. Lyngby, Denmark
2
The Danish National Research Foundation’s Center for Functionally Integrative Neuroscience
Aarhus University Hospital, Aarhus, Denmark
3
Department of Computer Science, University of Toronto, Toronto, Canada
4
Danish Research Centre for Magnetic Resonance, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark
5
Rotman Research Institute, Baycrest Centre for Geriatric Care, Toronto, Canada
6
Institute of Medical Science, University of Toronto, Toronto, Canada
7
Institute of Medical Biophysics, University of Toronto, Toronto, Canada
Keywords:
Neuroimaging, Classification, Multivariate Analysis, Model Interpretation, Model Visualization, Sensitivity
Map, NPAIRS Resampling, Functional Magnetic Resonance Imaging.
Abstract:
Classification models are becoming increasing popular tools in the analysis of neuroimaging data sets. Be-
sides obtaining good prediction accuracy, a competing goal is to interpret how the classifier works. From a
neuroscientific perspective, we are interested in the brain pattern reflecting the underlying neural encoding of
an experiment defining multiple brain states. In this relation there is a great desire for the researcher to gen-
erate brain maps, that highlight brain locations of importance to the classifiers decisions. Based on sensitivity
analysis, we develop further procedures for model visualization. Specifically we focus on the generation of
summary maps of a nonlinear classifier, that reveal how the classifier works in different parts of the input do-
main. Each of the maps includes sign information, unlike earlier related methods. The sign information allows
the researcher to assess in which direction the individual locations influence the classification. We illustrate
the visualization procedure on a real data from a simple functional magnetic resonance imaging experiment.
1 INTRODUCTION
1.1 Background
Interest in applying multivariate analysis techniques
to functional neuroimaging data is increasing, see
e.g., (Lautrup et al., 1994; Mørch et al., 1997; Strother
et al., 2002; Cox and Savoy, 2003; LaConte et al.,
2005; O’Toole et al., 2007). A comprehensive intro-
duction to classification methods in function magnetic
resonance imaging (fMRI) is provided in (Pereira
et al., 2009). Widely used classification schemes
include kernel methods such as support vector ma-
chines (SVMs) (Cox and Savoy, 2003; Davatzikos
et al., 2005; LaConte et al., 2005; Mour
˜
ao Miranda
et al., 2005). In kernel based learning, the input data
is implicitly mapped into a high-dimensional feature
space, and the classification model finds a linear de-
cision boundary in the feature space. Typical kernel
based learning methods are capable of constructing
arbitrary nonlinear decision boundaries in the input
space (i.e. the space of the measurements). For ad-
ditional discussion of nonlinear classification in neu-
roimaging, see (Mørch et al., 1997; Cox and Savoy,
2003; LaConte et al., 2005; Haynes and Rees, 2006;
Pereira et al., 2009; Misaki et al., 2010; Schmah
et al., 2010; Rasmussen et al., 2011). While linear
methods are of limited used when faced with nonlin-
ear data, nonlinear methods may require unavailable
large samples to generalize well (Mørch et al., 1997).
Another limitation that has hampered the application
of nonlinear kernel methods is the lack of well es-
254
M. Rasmussen P., Schmah T., H. Madsen K., E. Lund T., Yourganov G., C. Strother S. and K. Hansen L..
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN NEUROIMAGING - Signed Sensitivity Maps.
DOI: 10.5220/0003785602540263
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2012), pages 254-263
ISBN: 978-989-8425-89-8
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

tablished simple deterministic visualization schemes
(LaConte et al., 2005).
The aim of our present work is to develop further
procedures for interpretation/visualization of nonlin-
ear classification models.
1.2 Related Work
The sensitivity analysis that we investigate for model
interpretation is based on early work in (Zurada et al.,
1994; Zurada et al., 1997). (Kjems et al., 2002) have
developed probabilistic sensitivity maps as a generic
technique that can be applied to any model in order
to visualize the features with respect to their impor-
tance in classification. This sensitivity map technique
has been used for extraction of a single global sum-
mary map of the features importances to a trained
classifier. The procedure has been applied within the
field of neuroimaging in linear discriminant analysis
(Kjems et al., 2002), quadratic discriminant analysis
(Yourganov et al., 2010), and nonlinear kernel models
(Rasmussen et al., 2011), and in skin cancer detection
by Raman spectroscopy using neural networks (Sig-
urdsson et al., 2004).
For model interpretation (Baehrens et al., 2010)
recently proposed a general methodology for inter-
pretation of classifiers by exploring ”local explana-
tion vectors” that are defined as class probability gra-
dients. This procedure identifies features that are im-
portant for prediction at localized points in the data
space. (Golland et al., 2005) proposed a similar lo-
calized interpretation approach for support vector ma-
chine (SVMs) in the context of analysis of differ-
ences in anatomical shape between populations. They
aimed for a representation of the differences between
two classes captured by the classifier in the neighbor-
hood of data examples. These two procedures give a
localized visualization of the classifier, since they pro-
vide measures (here images) of each feature’s impor-
tance at particular points of interest in the data space.
1.3 Contribution of this Work
In the present work we focus on extending the sen-
sitivity map approach of (Kjems et al., 2002) to also
allow for extraction of summary maps with sign infor-
mation. Originally, this method of building sensitiv-
ity maps did not contain sign information: the voxel’s
weight reflected the the relative importance of a par-
ticular voxel to the classifiers decisions. However, it
is relevant to also consider maps containing sign in-
formation, which indicates whether a voxel’s signal
should be increased or decreased in order to increase
the probability of a particular data observation being
assigned to a specific class. The approach of (Kjems
et al., 2002) did not contain sign information due to
the difficulty of cancellation of opposite signs in an
average. In our approach, the cancellation problem is
mitigated to some extent by an unsupervised cluster-
ing step, by which we derive maps as weighted aver-
ages.
We aim for the characterization of trained classi-
fiers not by a single global summary map, but rather
a series of localized summary maps containing sign
information. The maps proposed in (Baehrens et al.,
2010; Golland et al., 2005) contain sign information,
and in principle these approaches provide one map per
data observation. In the present study we were inter-
ested in deriving maps containing sign information,
and also to extract only relatively few representative
maps in order to maintain simplicity of the model’s
visualization. This has potential to enhance the inter-
pretation of how trained classifiers function.
Using the NPAIRS resampling framework
(Strother et al., 2002) to assess the reliability/stability
of the extracted maps is a key element in the analysis
of our proposed procedure.
2 THEORY
In this section we briefly outline the concepts of su-
pervised learning, and review the basics of kernel
Fisher’s discriminant (KFD) analysis that we use for
classification. This is followed by an introduction to
the theoretical framework of the sensitivity map for
model visualization. The novel contribution in this
section is the discussion of four procedures for ob-
taining brain maps from a trained classifier, and in
particular one of these procedures based on cluster-
ing.
2.1 Classification Setup
We consider a multi-class problem, where we have
a labeled data set D = {x
n
,c
n
}
N
n=1
, where x is a P
dimensional input vector while c is the corresponding
class label that groups x into C disjoint classes.
Kernel based methods work implicitly in a fea-
ture space F that is related to the input space X
by a mapping φ : X → F , where φ(·) is a function
that returns a feature vector φ(x) corresponding to
an input point x. Rather than working in the feature
space kernel based methods work on a kernel func-
tion k(x
i
,x
j
) = φ(x
i
)
>
φ(x
j
) that returns inner prod-
ucts in feature space. Examples of kernels are the
linear kernel k(x
i
,x
j
) = x
>
i
x
j
and the RBF kernel
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN NEUROIMAGING - Signed Sensitivity Maps
255

k(x
i
,x
j
) = exp(||x
i
− x
j
||
2
/q). For a further introduc-
tion to kernel based learning we refer to the literature,
e.g. (Shawe-Taylor and Cristianini, 2004).
KFD analysis is a supervised dimensionality re-
duction technique (Mika et al., 1999), and a nonlin-
ear generalization of Fisher’s discriminant analysis.
KFD seeks to find optimal projection directions along
which the ratio of the between-class scatter to the to-
tal scatter is maximized. In the multi-class classifica-
tion problem the Fisher’s discriminant is given by the
matrix A, a C − 1 column matrix, that optimizes the
objective function
L(A) =
|A
>
S
B
A|
|A
>
(S
T
+ λI)A|
(1)
where S
B
=
∑
C
c=1
N
c
(m
c
− m)(m
c
− m)
>
is the
between-class scatter matrix, and S
T
=
∑
N
i=1
(φ(x
i
) −
m)(φ(x
i
)−m)
>
is the total scatter matrix, with N
c
de-
noting the number of samples in class c and m
c
and
m class means and grand mean respectively. Note
that we here consider regularized Fisher’s discrimi-
nant analysis, where λ is a regularization parameter
(Friedman, 1989). There exist several ways to solve
the above optimization problem. One is to consider
the following generalized eigenvalue problem (Zhang
et al., 2009b)
S
B
A = (S
T
+ λI)AD, (2)
where A holds the eigenvectors in the columns and
D holds the corresponding eigenvalues in the diag-
onal. Since the eigenvectors can be expressed as
A =
∑
N
n=1
(φ(x
i
) − m)b
>
i
, where b
i
is a coefficient
vector with C − 1 rows, we can reformulate eq. (2)
in terms of the kernel matrix
CWCB = (CC + λC)BD, (3)
where C = HKH is the centered kernel matrix K, with
the centering matrix defined as H = I
N
− 1
N
1
0
N
/N,
and W is an (N × N) positive symmetric weight ma-
trix with elements W
i, j
= 1/N
c
if x
i
and x
j
belongs to
the same class and W
i, j
= 0 otherwise. Based on the
solution to eq. (3) we can obtain the projection of a
feature space vector φ(x) onto A as
z = A
>
(φ(x) − m) = B
>
H(k
x
−
1
N
K1
N
), (4)
where k
x
= (k(x
1
,x),...,k(x
N
,x))
>
, and B is an (N ×
(C − 1)) matrix.
On top of the projection of data points onto the
subspace identified by KFD, we implement a simple
classifier assuming a Gaussian noise model. The like-
lihood function is
p(z
x
|µ
c
,σ
2
) = (2πσ
2
)
−
C−1
2
exp(−
1
2σ
2
||z
x
− µ
c
||
2
),
(5)
with µ
c
denoting the mean of projections of class c
and variance σ
2
shared across classes. Note that we
here use the notation z
x
to emphasize that z is a vector
valued function of x. Classification is then performed
according to Bayes’ rule
p(c|z
x
) =
p(z
x
|c)p(c)
∑
C
c
0
=1
p(z
x
|c
0
)p(c
0
)
. (6)
In the following we will also refer to p(c|z
x
) as the
classifier’s c’th output channel.
2.2 Model Visualization
2.2.1 General Definition
Sensitivity analysis is a simple measure of the rela-
tive importance of the different input features (vox-
els, in the present context) to the classifier. We follow
the approach on (Kjems et al., 2002; Strother et al.,
2002) and aim for a visualization of the relative im-
portance of the input data for a given function f (x) in
a stochastic environment with a distribution over the
inputs given by the probability density function p(x).
We define the signed sensitivity map by
s =
Z
A
∂ f (x)
∂x
p(x)dx, (7)
where s is a P dimensional vector, where the j’th
element holds the sensitivity measure corresponding
to the j’th voxel. Here A denotes the region of in-
tegration (some region of the image space). Hence,
the map summarizes the gradient field of the function
f (x) in some region A.
If f (·) is a nonlinear function, cancellation may
occur since some regions of the input space can have
positive sensitivity while other regions can have neg-
ative sensitivity. To avoid such cancellation we also
consider a map based on the squared sensitivities
s =
Z
A
∂ f (x)
∂x
2
p(x)dx. (8)
In practice the maps are approximated as a finite sum
over observations, for example eq. (7) is approxi-
mated by
s =
1
N
I
∑
n∈I
∂ f (x)
∂x
|
x=x
n
, (9)
where the sum is calculated based on a set I contain-
ing N
I
observations.
2.2.2 Choice of Visualization Function
Different choices for the visualization function f (x)
exist.
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
256

One choice is to use the posterior probability in
eq. (6)
f (z
x
) = p(c|z
x
), (10)
and this is the approach in (Baehrens et al., 2010).
A variant hereof is
f (z
x
) = log(p(c|z
x
)), (11)
that is the approach of (Kjems et al., 2002). In the
present paper we follow this approach and use eq.
(11) as a visualization function.
Another possibility is to use the decision func-
tion(s) as a visualization function. This is the ap-
proach in (Yourganov et al., 2010; Rasmussen et al.,
2011). For example, if we build a nearest mean classi-
fier on top of the subspace identified by KDF we have
C output channels where the c’th channel is given by
f (z
x
) = ||z
x
− µ
c
||
2
. (12)
2.2.3 Choice of Summation Region and Output
Channel(s)
The set I over which the summation in eq. (9) is done
and one or more output channels must be selected in
order to construct the map. Many possible variations
exist, and in the following we will outline just a few.
Procedure I
Let all observations be members of I. Furthermore
we consider all output channels, and hence obtain a
grand average map as
s
ga
=
1
C
C
∑
c=1
1
N
I
∑
n∈I
∂log(p(c|z
x
))
∂x
|
x=x
n
2
, (13)
where we square the single sensitivities to avoid
potential cancellations.
Procedure II
Let all observations in a class c be members of I
c
and
let only members of I
c
contribute to the sum over
output channel c. We obtain a grand average map as
s
ga
=
1
C
C
∑
c=1
1
N
I
c
∑
n∈I
c
∂log(p(c|z
x
))
∂x
|
x=x
n
2
, (14)
which is the procedure in (Kjems et al., 2002).
Procedure III
Here we are interested in a map reflecting an inter-
class contrast. Let all observations in a class c
0
be
members of I
c
0
and consider an output channel c. We
obtain a map as
s
c|c
0
=
1
N
I
c
0
∑
n∈I
c
0
∂log(p(c|z
x
))
∂x
|
x=x
n
, (15)
where we use the notation s
c|c
0
to indicate that we
consider the sensitivity with respect to output channel
c of the classifier, and the summation is performed
over the members class c
0
. This map describes how
observations in an input space region characterized by
the members in class c
0
should be changed in general
in order to increase the posterior probability of class
c. Note that this procedure can be problematic if the
derivatives have different sign across the members
in c
0
. One way to deal with this issue is to use the
definition eq. (8) to obtain a sensitivity map with no
sign information.
Procedure IV
If there exists a considerable heterogeneity in the
sign of the gradients within class c
0
, as discussed
above, we propose the following as an alternative to
using the squared gradients in order to derive signed
sensitivity maps: We consider specific classes c and
c
0
, as in the Procedure III, with I
c
0
comprised of all
observations in c
0
. We then perform a soft clustering
of all pairs (x
n
,s(x
n
)) for n ∈ I
c
0
, where s(x
n
) equals
∂log p(c|z
x
)/∂x
|
x=x
n
. Hence, we are interested in
patterns based on observations that are similar both
with respect to their input space location and their
sensitivity measure. The soft clustering takes the
form of a Gaussian mixture model (GMM) with the
number of clusters optimized by cross-validation. Fi-
nally, we construct a sensitivity map for each cluster,
by weighting the sensitivities s(x
n
) by weights w
k
n
defined to be the posterior probability of (x
n
,s(x
n
))
being in the given cluster k
s
c|c
0
,k
=
1
N
I
c
0
∑
n∈I
c
0
w
k
n
∂ f (x)
∂x
|
x=x
n
. (16)
If we assume that p(c|z
x
) is smooth, then taking a
large enough number of clusters will result in the vec-
tors s(x) within each cluster being similar, which will
reduce the sign cancellation problem and also obscure
less structure.
Specifically we estimate the weights in eq. (16) as
follows: (1) For each of the observations x we con-
struct a feature vector f by stacking the correspond-
ing single sensitivity s and the observation x itself
f = [x; s] (x and s were both scaled to unit norm in
order to put them on the somewhat same scale), so
that f is a 2P-dimensional vector. (2) We perform
principal component analysis (PCA) and project the
feature vectors f onto a PCA subspace. (3) Based on
the low dimensional feature representation, we build
a Gaussian mixture model (GMM). To estimate the
number of components/clusters k we use cross vali-
dation, where we estimate the generalization error of
the GMM by evaluating the likelihood function on the
left out fold.
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN NEUROIMAGING - Signed Sensitivity Maps
257

3 MATERIALS & METHODS
This section provides a description of the fMRI data
set used for illustration. This is followed by a de-
scription of our classification setup and the resam-
pling scheme used for model evaluation.
3.1 Functional MRI Data Set - Visual
Paradigm
Six healthy subjects were enrolled after informed con-
sent as approved by the local Ethics Committee. The
fMRI data set was acquired on a 3T (Siemens Mag-
netom Trio) scanner using a 8-channel head coil (In-
vivo, Florida, USA). The participants were subjected
to four visual conditions presented on a screen with
the following sequence: (1) no visual stimulation
(no), (2) reversing checkerboard on the left half of the
screen (left), (3) reversing checkerboard on the right
half of the screen (right), (4) reversing checkerboard
on both halfs of the screen (both). Each stimulus con-
dition was presented for 15 seconds followed by 5.04
seconds of rest with no visual stimulation. The stim-
ulation sequence was repeated 12 times in the exper-
imental run, and 576 scan volumes were acquired in
total.
Pre-processing of the fMRI time series was
conducted using the SPM8 software package
(http://www.fil.ion.ucl.ac.uk/spm) and comprised
the following steps for each subject: (1) Rigid body
realignment of the EPI images to the mean volume in
the time series. (2) Spatial normalization of the mean
EPI image to the EPI template in SPM8. (3) The
estimated warp field was applied to the individual EPI
images. The normalized images were written with 3
mm isotropic voxels. (4) For visualization purposes
the anatomical scan was spatially normalized to the
T1 template in SPM8, using the same settings as for
the EPI images. (5) The EPI images were smoothed
with an isotropic 8 mm full-width half-maximum
Gaussian kernel. (6) The data were masked with
a rough whole-brain mask (75257 voxels). (7) To
remove low frequency components from the time
series, a set of discrete cosine basis functions up to a
cut-off period of 128 seconds were projected out of
the data. (8) Within each subject the individual voxel
time series were standardized (each voxel subtracted
the mean and scaled by the standard deviation of
the voxel’s time series). Further details on the
data acquisition and pre-processing are provided in
(Rasmussen et al., 2011).
Figure 1 shows the average fMRI images for each
of the four experimental conditions, and is shown in
order to assist the reader in interpreting the subse-
(no)
(left)
(right)
(both)
-8
7 22 37
Figure 1: Average EPI images across all six subjects for the
four conditions in the fMRI data set. The data was standard-
ized within each subject. Warm and cold colors are positive
and negative values respectively. The maps are thresholded
to show the upper 10 percentile of the voxel absolute value
distribution, projected onto an average structural scan of the
six subjects included in the analysis. Numbers under the
slices denote z coordinates in MNI space. Slices are dis-
played according to neurological convention (right side of a
brain slice is the right side of the brain).
quent figures. Negative signal occurs due to the stan-
dardization of data within each subject. The maps
are thresholded to show the upper 10 percentile of the
voxel absolute value distribution (as in all subsequent
maps shown).
3.2 Classification Setup and Model
Evaluation
In the analysis of the fMRI data set, we considered
subjects as the basic resampling unit, where the clas-
sifier was trained on data from a subset of subjects,
while the target labels were inferred for scans from
subjects in the out-of-sample subset. Scans 7-11 in
each epoch were used in the predictive modeling, and
the remaining volumes were discarded to avoid con-
taminating effects of the BOLD signal. We aimed for
a ”whole-brain” and single block classification with
temporal averaging of scans within the same block.
No feature selection prior to the application of the
classification models was performed. Two binary
classification task were formulated:
• Classification task I: We considered a four class
classification task, where scans from the four con-
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
258

ditions {no, left, right, both} were assigned to the
classes {1,2,3,4}. We expect observations belong-
ing to the same class to be relatively homogeneous
in this classification task.
• Classification task II: scans from condition (no)
and (both) were assigned to class 1, while scans
from condition (left) and (right) were assigned to
class 2. By this labeling we intended to intro-
duce an artificial coupling between brain regions,
equivalent to computer science’s xor function. In
this classification task we expect a relatively large
degree of heterogeneity between observations be-
longing to that same class.
For classification we used KFD analysis with the
RBF kernel. The model estimation requires selec-
tion of the regularization parameter λ in KFD and
selection of the q parameter that controls the kernel
width of the RBF kernel. We let λ range over the
interval 2
−15
...2
30
relative to the average non-zero
eigenvalue of the data covariance matrix, while q was
varied in the range 2
−5
...2
16
relative to the average
input-space distance to the nearest 25% points across
all training examples.
For model evaluation we used the NPAIRS resam-
pling scheme (Strother et al., 2002). In this cross-
validation framework the data were split into two par-
titions of equal size (three subjects in each partition).
The model was trained on the first split and the predic-
tion accuracy was estimated from the second split and
vice versa, yielding two estimates of the prediction
accuracy. We measure the prediction accuracy by the
posterior probability for the correct class label. These
prediction accuracies were averaged and considered
as the prediction metric (p) of the NPAIRS scheme.
In addition we extracted a grand average sensitivity
map according to eq. (13) for each of the two classi-
fication models. The Pearson’s correlation coefficient
between spatial maps derived from the two models
was calculated as a spatial reproducibility metric (r).
Each map vector was scaled to unit norm, and the
scatter plot of the maps from each model was pro-
jected onto a signal axis and an uncorrelated noise
axis as described in (Strother et al., 2002). The pro-
jection onto the signal axis was scaled by the stan-
dard deviation of the noise projection, which gave a
reproducible statistical parametric image (rSPI). This
procedure was repeated for all possible splits of the
subjects (10 resampling runs).
We use the p and r metrics for model optimiza-
tion, where we choose model parameters that maxi-
mize both metrics jointly, in terms of the Euclidean
distance to the point (p,r)=(1,1) over the entire space
of cross-validated results (Strother et al., 2002; Zhang
et al., 2009a).
In order to interpret the classifier based on brain
maps containing sign information we also derived
maps according to Procedure III in section (2.2).
Furthermore, for classification task II we derived
maps according to Procedure IV in Section (2.2),
since we expected a relatively large heterogeneity be-
tween single sensitivities of observations within the
same class. We performed Steps 1-3 of Procedure
IV (Section (2.2)) within the NPAIRS resampling
scheme. For the PCA subspace in Step 2, we used
the space spanned by the first two PCs (this was a
heuristic choice). The most likely number of clus-
ters k ∈ [1,...,6] in the GMM was found by maxi-
mizing the mean likelihood over the 10 resampling
splits. We then performed a second pass through the
NPAIRS procedure, where we fixed k across all splits
and resampling runs in order to obtain the same num-
ber of clusters across models. For each split we fitted
a GMM and derived sensitivity maps according to eq.
(16). In order to derive rSPIs the clusters labels must
be aligned across splits. We used a simple reference
filtering procedure, where the cluster’s labels of a par-
ticular split was permuted in order to maximize the
correlation between sensitivity maps across splits.
4 RESULTS AND DISCUSSION
Figure 2 shows average rSPIs derived from the classi-
fication models in classification task I (four classes).
The average prediction accuracy was 0.9229 in terms
of average posterior probability of the correct class.
Figure 2(A) shows the average rSPI across NPAIRS
splits based on the grand average maps s
ga
derived ac-
cording to Procedure I in Section 2.2. The map iden-
tifies that voxels in the visual cortex contribute the
most with relevant information to the classifiers. The
average reproducibility of the map across splits was
0.81. Note that the rSPI only contains positive values
since it is based on squared sensitivities. Figure 2(B)
shows average rSPIs that are based on the interclass
contrast (Procedure III in Section 2.2). According to
the map s
le f t|no
a signal increase primarily in the right
visual cortex will increase the posterior probability of
the scans in class (no) being classified as belonging
to class (left). Figure 1 indicates that this is reason-
able, since the condition (left) is characterized by a
larger signal in the right visual cortex relative to the
(no) condition. Likewise, the map s
le f t|right
indicates
that lowering the signal in the left visual cortex and in-
creasing signal in the right visual cortex will increase
the posterior probability of the scans in class (right)
being classified as (left). The reproducibility of the
maps in Figure 2 are fairly high ranging from 0.69-
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN NEUROIMAGING - Signed Sensitivity Maps
259

-8
7 22 37
s
left∣no
0.81
0.80
0.78
0.69
(A)
(B)
s
left∣right
s
left∣both
s
ga
Figure 2: Classification task I with a four class classification
task. Different brain maps were extracted from the trained
classifier. Panel (A) shows an average rSPI based on the
grand average map (Procedure I in Section 2.2). Panel (B)
shows signed rSPIs based on Procedure III in Section 2.2.
The notation e.g. s
le f t|no
means that the map indicates how
scans in class ’no’ should be changed in order to increase
the posterior probability of class ’left’. The numbers right to
the brain slices denote the reproducibility estimated within
the NPAIRS resampling framework. The rSPIs are thresh-
olded to show the upper 10 percentile of the distribution of
the absolute voxel values, projected onto an average struc-
tural scan of the six subjects included in the analysis. Num-
bers under the slices denote z coordinates in MNI space.
Slices are displayed according to neurological convention.
0.80.
Figure 3(C-E) shows average rSPIs derived from
the classification models in classification task II (xor-
problem). The average prediction accuracy was
0.905. Figure 3(C) shows the average rSPI across
NPAIRS splits based on the grand average maps s
ga
derived according to Procedure I in Section 2.2. The
maps identify that voxels in the visual cortex that con-
tribute the most relevant information to the classifiers.
The average reproducibility of the maps across splits
was 0.81. Note that Figure 3(C) resembles Figure
2(A), which means that the regions identified as being
relevant to classification in tasks I and II are similar.
Figure 3(D) shows average rSPIs based on interclass
contrasts (Procedure III in Section 2.2). Due to the
potential heterogeneity within the classes, the sensi-
tivity maps were based on squared single sensitivities.
Hence, no sign information is present in the maps.
For comparison, we derived signed class specific
sensitivities directly as in classification task I Figure
2(B) (without squaring), and found that the repro-
ducibilities of the maps were reduced to 0.36. This
decrease in reproducibility is presumably due to can-
cellation effects.
Figure 3(E) shows the rSPIs obtained from Pro-
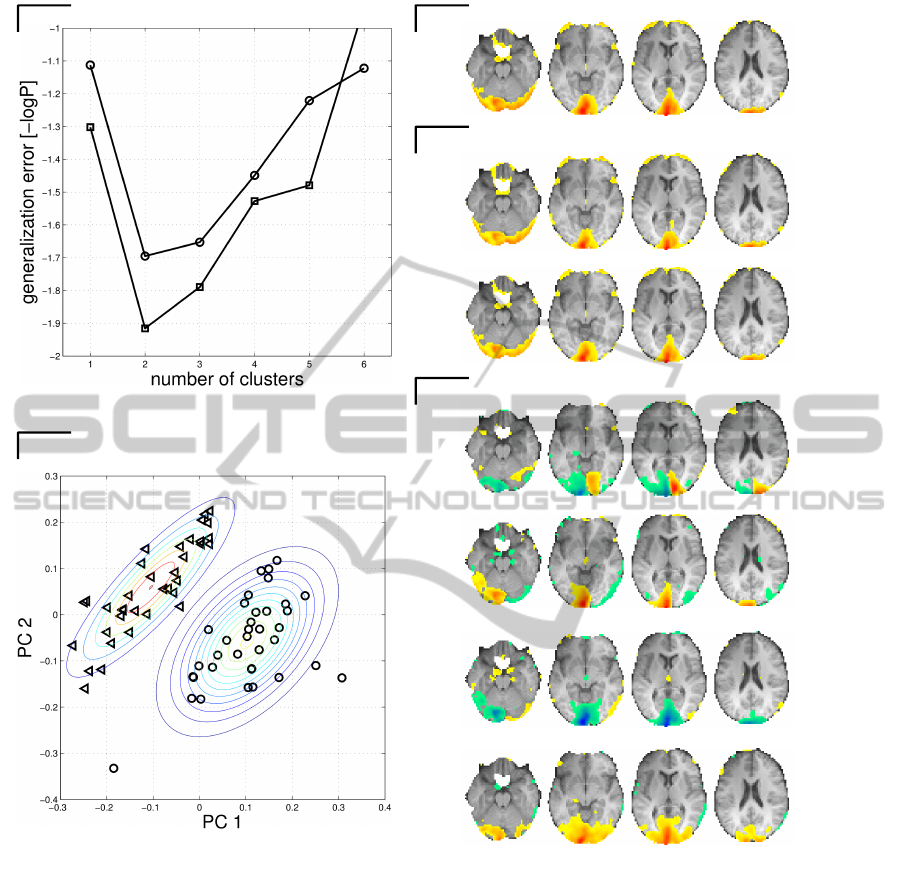
cedure IV in Section 2.2. The first and second rows
of Figure 3(E) can be seen as refinements of the first
row of Figure 3(D), obtained by dividing the class
{le f t,right} into two clusters. Similarly, the third and
fourth rows of Figure 3(E) are refinements of the sec-
ond row of Figure 3(E).
The clustering process is illustrated in Figure 3(A-
B). Figure 3(B) shows an example of clustering of
observations within class {le f t,right}. In both class
{no,both} and class {le f t,right} we found evidence
of two clusters based on generalization error mea-
sured as the negative likelihood of the Gaussian mix-
ture model (Figure 3(A)). Based on the cluster anal-
ysis we derive two maps for each of the two classes
according to Procedure IV in Section 2.2. By this pro-
cedure we obtained maps with sign information. Fig-
ure 3(E) shows the resulting four rSPI (two for each
class). For example s
{no,both}|{le f t,right},cluster1
denotes
that the sensitivity map is based on the output class
{no,both}, and the sum in eq. (16) is calculated over
the members of class {le f t,right}, where the weight-
ing factor w
k
n
is based on the posterior probability
for observations belonging to cluster 1. The repro-
ducibility values of the sensitivity maps are moderate,
ranging from 0.53-0.61. Note that these reproducibil-
ity values are lower than for Procedure III based
on squared single sensitivities (see Figure 3(D)), but
higher than for Procedure III with signed sensitivities
(mean reproducibility 0.36, as stated earlier). To fur-
ther interpret the maps in Figure 3(E) we calculated
the average weight factor w
k
n
in eq. (16) for each of the
sub clusters. For the map s
{no,both}|{le f t,right},cluster1
we found that the members of condition (left) had
an average weighting factor of 0.0038 in the sum,
while the members of the condition (right) had an av-
erage weighting factor of 0.9857. Hence, members of
the condition (right) contribute the most to this map.
Likewise, the members of condition (left) contributed
the most to the map s
{no,both}|{le f t,right},cluster2
. For
the map s
{le f t,right}|{no,both},cluster1
we found that the
members of condition (no) had an average weighting
factor of 0.0000 in the sum, while the members of
the condition (both) had an average weighting factor
of 0.9986. Hence, members of the condition (both)
contribute the most to this map. Likewise, the mem-
bers of condition (no) contribute the most to the map
s
{le f t,right}|{no,both},cluster2
.
Note the similarity of the maps in the first row of
Figure 3(E) (s
{no,both}|{le f t,right},cluster1
, where cluster
1 corresponds to class right, as just explained) and the
second row of Figure 2(B) (s
le f t|right
). Also the fourth
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
260
-8
7 22 37
s
ga
0.81
0.78
0.76
0.53
0.56
0.61
0.56
(C)
(D)
(E)
s
{no,both}∣{left,right}
s
{no,both}∣{left,right}
s
{no,both}∣{left,right},cluster 2
s
{no,both}∣{left,right},cluster 1
s
{left,right}∣{no,both},cluster 1
s
{left,right}∣{no,both},cluster 2
(A)
(B)
Figure 3: Classification task II. Panel A-B: Structure of single sensitivities. See Procedure IV in Section 2.2 for further
details. Modeling of the data distribution was based on a Gaussian mixture model (GMM). Panel A shows the generalization
error as a function of the number of components (averaged over 10 resampling runs) in the GMM. The generalization error
was calculated by evaluation of the likelihood function of the GMM based on four-fold cross validation performed as a nested
loop within each NPAIRS split half. The circles corresponds to the class s
{no,both}|{le f t,right}
, and the squares corresponds
to the s
{le f t,right}|{no,both}
single sensitivity measures. In both classes we observe evidence for two clusters/components.
Panel B shows contours of the probability densities of a GMM fitted to s
{no,both}|{le f t,right}
sensitivities of a single split
of the data. The triangles corresponds to the observations in condition (left) and the circles corresponds to observations in
the condition (right). Note that this labeling was not ”visible” to the modeling procedure. Panel C-E: Interpretation of the
trained classifier. Different brain maps were extracted from the classifier. Panel (C) shows an average rSPI based on the
grand average sensitivity map (Procedure I in Section 2.2). Panel (D) shows rSPIs based on single class sensitivities. The
notation eg. s
{no,both}|{le f t,right}
means that the map indicates how scans in class {le f t,right} should be changed in order to
increase the posterior probability of class {no,both}. Note that the maps does not contain sign information (Procedure III in
Section 2.2). Panel (E) shows rSPIs based on single class sensitivity, where each class is characterized by two sub-clusters
(Procedure IV in Section 2.2). The numbers right to the brain slices denote the reproducibility estimated within the NPAIRS
resampling framework. The rSPIs are thresholded to show the upper 10 percentile of the distribution of the absolute voxel
values, projected onto an average structural scan of the six subjects included in the analysis. Numbers under the slices denote
z coordinates in MNI space. Slices are displayed according to neurological convention.
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN NEUROIMAGING - Signed Sensitivity Maps
261

row of Figure 3(E) ((s
{le f t,right}|{no,both},cluster2
, where
cluster 2 corresponds to class no) is similar to the first
row of Figure 2(B) (s
le f t|both
); and the third row of
Figure 3(E) (s
{le f t,right}|{no,both},cluster1
, where cluster
1 corresponds to class both) is similar to the third row
of Figure 2(C) (s
le f t|both
). The significance of these
observations is that Procedure IV has succeeded in
providing similar information when applied to Clas-
sification Task II (the 2-class xor-problem) as the in-
formation given by Procedure III applied to Classifi-
cation Task I (the 4-class classification), even though
the available class labels in Task II are less informa-
tive and the two classes no, both and left, right are
heterogeneous. This suggests that Procedure IV can
provide useful information about the nature of non-
linear classifiers when applied to complex, heteroge-
neous classes.
5 CONCLUSIONS
The established probabilistic sensitivity map proce-
dure provides a global summary map of the relative
importance of voxels to a trained classifier (Kjems
et al., 2002). However, no sign information is present
in such a map. In the present work we have pro-
posed a procedure to allow for generation of sum-
mary maps with sign information. Furthermore, we
have proposed a clustering procedure that is applica-
ble in cases where relatively large heterogeneity be-
tween observation exists which may degrade the per-
formance of the model visualization due to cancella-
tion effects.
As a proof of concept, we have illustrated the ap-
proach on a data set from a simple fMRI experiment,
with classes deliberately defined to be heterogeneous.
Our procedure successfully recovered known struc-
ture in the classes. We also found that the maps pro-
duced for this data set are robust, in the sense that
they are reproducible as judged by the NPAIRS re-
sampling framework. We showed that reproducibility
is improved by the new clustering procedure.
Our results suggest that our new method of model
visualization may be useful in visualizing nonlinear
classifiers trained on heterogeneous classes. Further
work is needed to compare variations of the method,
in particular different possible choices of the visual-
ization function (see Section 2.2.2), and to validate
the method on a larger variety of real or synthetic data.
ACKNOWLEDGEMENTS
This work is partly supported by the Danish Lundbeck
Foundation through the program www.cimbi.org. The
Simon Spies Foundation is acknowledged for dona-
tion of the Siemens Trio scanner. Kristoffer H. Mad-
sen was supported by the Danish Medical Research
Council (grant no. 09-072163) and the Lundbeck
Foundation (grant no. R48-A4846).
REFERENCES
Baehrens, D., Schroeter, T., Harmeling, S., Kawanab, M.,
Hansen, K., and M
¨
uller, K.-R. (2010). How to ex-
plain individual classification decisions. Journal of
Machine Learning Research, 11:1803–1831.
Cox, D. D. and Savoy, R. L. (2003). Functional magnetic
resonance imaging (fMRI) ”brain reading”: detecting
and classifying distributed patterns of fMRI activity in
human visual cortex. NeuroImage 19, pages 261–270.
Davatzikos, C., Ruparel, K., Fan, Y., Shen, D., Acharyya,
M., Loughead, J., Gur, R., and Langleben, D. (2005).
Classifying spatial patterns of brain activity with ma-
chine learning methods: application to lie detection.
NeuroImage, 28(3):663–668.
Friedman, J. H. (1989). Regularized discriminant analysis.
J. Am. Statistical Assoc., 84:165 – 175.
Golland, P., Grimson, W. E. L., Shenton, M. E., and Kikinis,
R. (2005). Detection and analysis of statistical differ-
ences in anatomical shape. Medical Image Analysis,
9:69–86.
Haynes, J. D. and Rees, G. (2006). Decoding mental states
from brain activity in humans. Nature Reviews Neu-
roscience, 7(7):523–534.
Kjems, U., Hansen, L. K., Anderson, J., Frutiger, S., Mu-
ley, S., Sidtis, J., Rottenberg, D., and Strother, S. C.
(2002). The quantitative evaluation of functional neu-
roimaging experiments: mutual information learning
curves. NeuroImage, 15(4):772–786.
LaConte, S., Strother, S., Cherkassky, V., Anderson, J., and
Hu, X. (2005). Support vector machines for temporal
classification of block design fMRI data. NeuroImage,
26:317–329.
Lautrup, B., Hansen, L., Law, I., Mørch, N., Svarer, C.,
and Strother, S. (1994). Massive weight sharing: A
cure for extremely ill-posed problems. Proceedings of
the Workshop on Supercomputing in Brain Research:
From Tomography to Neural Networks. World Scien-
tific, Ulich, Germany, pages 137–148.
Mika, S., R
¨
atsch, G., Sch
¨
olkopf, B., Smola, A., Weston, J.,
and M
¨
uller, K.-R. (1999). Invariant feature extraction
and classification in kernel spaces. Advances in Neu-
ral Information Processing Systems, 12:526–532.
Misaki, M., Kim, Y., Bandettini, P., and Kriegeskorte, N.
(2010). Comparison of multivariate classifiers and re-
sponse normalizations for pattern-information fMRI.
NeuroImage, 53(1):103–118.
Mørch, N., Hansen, L. K., Strother, S. C., Svarer, C., Rot-
tenberg, D. A., Lautrup, B., Savoy, R., and Paulson,
O. B. (1997). Nonlinear versus Linear Models in
Functional Neuroimaging: Learning Curves and Gen-
eralization Crossover. IPMI ’97: Proceedings of the
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
262

15th International Conference on Information Pro-
cessing in Medical Imaging, pages 259–270.
Mour
˜
ao Miranda, J., Bokde, A., Born, C., Hampel, H., and
M (2005). Classifying brain states and determining
the discriminating activation patterns: support vector
machine on functional MRI data. NeuroImage, 28:980
– 995.
O’Toole, A. J., Jiang, F., Abdi, H., P Nard, N., Dunlop, J. P.,
and Parent, M. A. (2007). Theoretical, statistical, and
practical perspectives on pattern-based classification
approaches to the analysis of functional neuroimaging
data. Journal of Cognitive Neuroscience, 19:1735–
1752.
Pereira, F., Mitchell, T., and Botvinick, M. (2009). Machine
learning classifiers and fMRI: a tutorial overview.
NeuroImage, 45(1):S199–S209.
Rasmussen, P. M., Madsen, K. H., Lund, T. E., and Hansen,
L. K. (2011). Visualization of nonlinear kernel mod-
els in neuroimaging by sensitivity maps. NeuroImage,
55(3):1120 – 1131.
Schmah, T., Yourganov, G., Zemel, R., Hinton, G., Small,
S., and Strother, S. (2010). A Comparison of Classifi-
cation Methods for Longitudinal fMRI Studies. Neu-
ral Computation, 22:2729–2762.
Shawe-Taylor, J. and Cristianini, N. (2004). Kernel methods
for pattern analysis. Cambridge.
Sigurdsson, S., Philipsen, P., Hansen, L., Larsen, J.,
Gniadecka, M., and Wulf, H. (2004). Detection
of skin cancer by classification of Raman spec-
tra. IEEE Transactions on Biomedical Engineering,
51(10):1784–1793.
Strother, S., Anderson, J., Hansen, L., Kjems, U., Kustra,
R., Sidtis, J., Frutiger, S., Muley, S., LaConte, S., and
Rottenberg, D. (2002). The quantitative evaluation of
functional neuroimaging experiments: The NPAIRS
data analysis framework. NeuroImage, 15(4):747–
771.
Yourganov, G., Schmah, T., Small, S., PM, R., and Strother,
S. (2010). Functional connectivity metrics during
stroke recovery. Arch Ital Biol., 148(3):259–270.
Zhang, J., Anderson, J. R., Liang, L., Pulapura, S. K.,
Gatewood, L., Rottenberg, D. A., and Strother,
S. C. (2009a). Evaluation and optimization of fMRI
single-subject processing pipelines with NPAIRS and
second-level CVA. Magn Reson Imaging, 27:264–
278.
Zhang, Z., Dai, G., and Jordan, M. I. (2009b). A flexible
and efficient algorithm for regularized fisher discrim-
inant analysis. In ECML PKDD ’09: Proceedings of
the European Conference on Machine Learning and
Knowledge Discovery in Databases, pages 632–647,
Berlin, Heidelberg. Springer-Verlag.
Zurada, J., Malinowski, A., and Cloete, I. (1994). Sensitiv-
ity analysis for minimization of input data dimension
forfeedforward neural network. 1994 IEEE Interna-
tional Symposium on Circuits and Systems, 1994. IS-
CAS’94., 6:447–450.
Zurada, J., Malinowski, A., and Usui, S. (1997). Perturba-
tion method for deleting redundant inputs of percep-
tron networks. Neurocomputing, 14(2):177–193.
APPENDIX
In the following we show how to calculate the deriva-
tive used in eq. (7,8,9). First we calculate the deriva-
tive in eq. (4)
∂z
x
∂x
=
∂k
x
∂x
HB. (17)
We then calculate the derivative of the visualization
function
∂log(p(c|z
x
))
∂x
=
∂log(p(z
x
|c)P(c))
∂x
−
∂log(
∑
C
c
0
=1
p(z
x
|c
0
)P(c
0
))
∂x
=
−
∂||z
x
− µ
c
||
2
∂x
+
C
∑
c
0
=1
p(c
0
|z
x
)
∂||z
x
− µ
c
0
||
2
∂x
=
−
∂z
x
∂x
(z
x
− µ
c
) +
∂z
x
∂x
C
∑
c
0
=1
p(c
0
|z
x
)(z
x
− µ
c
0
) =
−
∂z
x
∂x
(z
x
− µ
c
) −
C
∑
c
0
=1
p(c
0
|z
x
)(z
x
− µ
c
0
)
!
=
−
∂k
x
∂x
HB
(z
x
− µ
c
) −
C
∑
c
0
=1
p(c
0
|z
x
)(z
x
− µ
c
0
)
!
. (18)
For example, for the linear kernel we have
∂k
x
∂x
= X, (19)
where X holds training observations in the columns.
For the RBF kernel we have
∂k
x
∂x
= MD
k
, (20)
where M in a (P × N) matrix that holds the elements
M
i, j
= x
j
i
− x
i
, with x
j
i
referring to the i’th element in
the j’th training example. D
k
is an (N × N) diagonal
matrix with the elements of k
x
in the diagonal.
VISUALIZATION OF NONLINEAR CLASSIFICATION MODELS IN NEUROIMAGING - Signed Sensitivity Maps
263
