
A NOVEL GAUSSIAN FITTING APPROACH FOR 2D GEL
ELECTROPHORESIS SATURATED PROTEIN SPOTS
Massimo Natale
1,2
, Alfonso Caiazzo
3
, Enrico M. Bucci
2,4
and Elisa Ficarra
1
1
Department of Control and Computer Engineering, Politecnico di Torino, Torino, Italy
2
BioDigitalValley srl, via Carlo Viola 78, Pont Saint Martin (AO), Italy
3
WIAS Berlin, Mohrenstrasse 39, 10997, Berlin, Germany
4
Istituto di Biostrutture e Bioimmagini, Via Mezzocannone 16, 80134, Naples, Italy
Keywords: Image analysis, Bi-dimensional electrophoresis, Proteomics, Software tools.
Abstract: Analysis of 2D-GE images is a hot topic in bioinformatics research, since currently available commercial
and academic software has proven to be not really effective and not completely automatic, often requiring
manual revision of spots detection and refinement of computer generated matches. In this work, we present
an effective technique for the detection and the reconstruction of over-saturated protein spots.
Firstly, it reveals overexposed areas where spots may be truncated, and plateau regions caused by smeared
and overlapped spots. As next, the correct distribution of pixel values in the overexposed areas and plateau
regions is recovered by a two-dimensional fitting based on a generalized Gaussian distribution approximat-
ing the spots volume. Pixel correction according to the generalized Gaussian curve in saturated and smeared
spots allows more accurate quantifications, providing more reliable image analysis results.
As validation, we process highly exposed 2D-GE image, containing saturate spots, with respect to the corre-
sponding non-saturated image, confirming that the method can effectively fix the saturated spots and enable
correct spots quantification.
1 INTRODUCTION
In the post-genomic era, two-dimensional gel elec-
trophoresis (2D-GE) is a powerful and widely used
method for the analysis of complex protein mixtures
extracted from cells, tissues, or other biological
samples. In 2D-GE proteins are separated according
to their charge (pI) by isoelectric focusing in the first
dimension, and according to their molecular weight
(MW) by SDS-PAGE in the second dimension. Each
resulting 2D-GE contains a few hundred up to sev-
eral thousands of protein spots whose volume corre-
lated with the protein expression in the sample. The
main goal of comparative proteomics is to match
protein spots between gels and define differences in
the expression level of proteins at different biologi-
cal states using image analysis software.
Good image capture is critical to guarantee opti-
mal performance of automated image analysis pack-
ages and generate reliable scientific data. It should
allow for detection from very low to high abundant
protein amounts. Through digitalization, a gel is
represented by a two-dimensional matrix of squares
or pixels. Each pixel of the generated image is de-
fined by its coordinates (x,y) and by its signal inten-
sity I encoded as greyscale level. The spatial coordi-
nates are defined by image resolution, while the
signal intensity is defined by dynamic range.
Saturation occurs when grey levels exceed the
maximum representable. When a spot becomes satu-
rated, the spot appears truncated, and any differences
in high pixel intensities cannot be resolved. No reli-
able quantitative data will be generated from a satu-
rated spot, and different authors recommend to
manually deleting the saturated spots, before analyz-
ing the gels with the available software (Berth,
2007).
Where different experimental states are being
compared the inclusion into the analysis of saturated
spots have the potential to bias normalization, in
particular if they have a variance that is a significant
proportion of the total spot volume (Miller, 2006).
Currently available commercial software (as
Delta2D, ImageMaster, PDQuest, Progenesis) are
not able to deal with specific protein spot distortions
found in the gel images (Maurer, 2006). Automatic
335
Natale M., Caiazzo A., M. Bucci E. and Ficarra E..
A NOVEL GAUSSIAN FITTING APPROACH FOR 2D GEL ELECTROPHORESIS SATURATED PROTEIN SPOTS.
DOI: 10.5220/0003789803350338
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2012), pages 335-338
ISBN: 978-989-8425-90-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

reconstruction of over saturated protein spots is not
possible using available protein analysis software.
The only option suggested by the producers of soft-
ware for 2D-GE analysis is to rescan the gel decreas-
ing the exposure (Nonlinear, 2011). Most of the
time, this is not possible because gel staining are
photosensitive, and loss intensity in few minutes.
Moreover, an image acquired with the largest
exposure also contains the highest number of spots
and thus also the lower-abundance protein spots.
Thus, decreasing the exposure causes the lower-
abundance protein spots to be neglected.
In this work, we present a novel algorithm for the
detection and the reconstruction of over-saturated
protein spots. Firstly, the algorithm reveals overex-
posed areas where spots may be truncated. Secondly,
this method reconstructs the over-saturated protein
spots through a Gaussian mathematical model that
simulates the distribution of the greyscale values in
the saturated spots. Furthermore, the algorithm will
be validated processing highly exposed 2D-GE im-
age, containing saturate spots, to the corresponding
non-saturated image.
2 METHODS AND RESULTS
2.1 Despeckle
After image scanning the 2D-GE are noisy, with
artifacts as scratches, air bubbles, and spikes.
Figure 1: A) Portion of 2D-GE gel where the spikes are
marked with black arrows. B) Portion of 2D-GE gel after
the 3x3 median filter. C) , D) 3D view of the portion of
gels
The aim of reducing speckles in a 2D-GE image
is to remove the noise without introducing any dis-
tortion in quantitative spot volume data.
We evaluated the filter size from 3x3 to 7x7. We
chose a 3x3 filter because it is effective in removing
most of the spikes with minor smoothing effects that
reduce the accuracy in the spot volume computation.
The result of 3x3 median filter is shown in figure 1.
In figure 1A is shown a portion of gel where the
spikes are marked with black arrows. In Figure 1C is
shown the portion of gel in 3D view where a spike is
marked with a red arrow. In Figure 1B and 1D are
shown the 2D-GE image and the 3D view after ap-
plying the 3x3 median filter.
2.2 Find Plateau Regions
The commercial software are able to detect a satu-
rated area when a region of the image reaches the
maximum value of greyscale. However, in most
cases a saturated area looks like a plateau zone,
where the pixels have similar intensity, but they do
not reach the maximum value of greyscale.
In these cases the commercial software not only
are unable to correct this aberration but they also fail
to detect it, inducing the operator to underestimate
the problem.
In order to identify plateau regions, we imple-
mented a morphological filter inspired by the Roll-
ing Ball algorithm (Sternberg, 1983). We designed
structural elements as ball of circular shape with
radius (RD) and height defined by greyscale value
tolerance (GVT). The RD represents the number of
pixels of the circle radius (Figure 2A). The GVT
represents the rate of gray values of the pixel in the
centre of the structural elements (Figure 2B).
Figure 2: A) The radius defines the pixels that are included
into the structural element. B) The greyscale value toler-
ance defines the variation within the gray values are con-
sidered to plateau.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
336

Figure 3: Saturation zone detected from our algorithm.
The figure shows the same gel acquired with three differ-
ent exposure. 2D-GE gel acquired with A) high, B) me-
dium, and C) low exposure.
The RD and the GVT are defined by a single pa-
rameter so that if the user sets, for example, 10 as
parameter, the RD will set of 10 pixels and the GVT
of the 10% of gray values of the pixel in (x,y) posi-
tion. The centre of the structural element is moved
along each pixel across the image. For each point the
maximum and minimum gray value within the given
RD is calculated. When the difference between
maximum and minimum is less than the GVT, the
area defined by local operator is considered as a
plateau area.
In order to test our procedure, we ran the method
on more than 50 2D-GE images, each acquired at 3
different expositions. An example is shown in fig-
ures 3A, 3B and 3C. These gels provide us the op-
portunity to see how the gray values are distributed
in the same spot. The plateau areas found by our
algorithm are filled in red.
Figure 3A contains a larger number of spots, but
some of them are saturated. In this case, only image
in figure 3C could be correctly analyzed by available
software. The other two images would be discarded
because of the large saturated areas that prevent
accurate protein expression measures. However,
researchers would be often interested in analyzing
the image acquired with the largest exposure (figure
3A), which also contains the highest number of spots
(and thus also the lower-abundance protein spots).
For this purpose we developed a refined algorithm
capable of calculating the original non-saturated
distribution of the gray values within the saturated
zone of the spot.
2.3 Gaussian Fitting
To determine the unknown distribution of gray val-
ues in the saturated area, we firstly assumed that the
intensity values distribution in the spot is described
by a Gaussian function of the form (1):
(
,,
)
=
√
2
exp −
2σ
(1)
where σ and M are, respectively, the standard devia-
tion and the average of intensity values, while x is a
pixel coordinate in the image. To determine σ and
M, we search the optimal fitting of the Gaussian
function in (1) with the values of the unsaturated
spot. In other words, given a set of m points with
coordinates (x
j
,y
j
), for j = 1, ..., m, the problem is to
find the couple of parameters (σ,M), such that the
differences (2)
f(x
j
, σ, M) - I
j
(2)
are small, where I is the pixel intensity. Choosing
the approximation criterion is equivalent to define an
error function, describing how far is the Gaussian
approximation from the original dataset in the unsa-
turated region. As error function we selected the sum
of the squared differences:
(
,
)
=(
(
,,)−
)
(3)
Other possibilities consist in taking different powers
of the differences, or including a set of weights, e.g.
to give different relevance to the point closer to the
saturation.
However, once the error function is chosen, the
problem results in finding the couple (σ,M) that
minimize the error function itself. A possible ap-
proach is to perform an exhaustive search on (σ,M)
values.
Figure 4: A) Portion of 2D-GE where all spots are cor-
rectly acquired. B) Portion of 2D-GE where two spots are
saturated. C) Plot profile of gray value found along the
green line as shown in 4A). D) Plot profile of gray value
found along the yellow line as shown in 4B). In red are
shown the saturated zone.
However, if the size of the parameters space is
too big, a more effective Newton-Raphson algorithm
to find the zero of the gradient can be employed. In
figure 4 we show the distribution of gray values in
non saturated spots (figure 4A) and in saturated
spots (figure 4B).The figure 4C and 4D show the
A NOVEL GAUSSIAN FITTING APPROACH FOR 2D GEL ELECTROPHORESIS SATURATED PROTEIN SPOTS
337
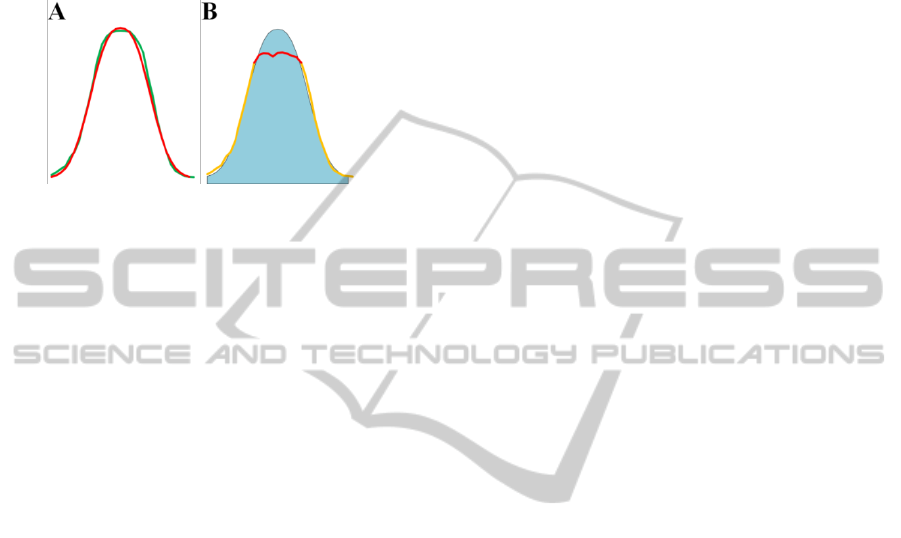
intensity distribution profiles. In figure 4B and 4D
are underlined the saturated value by red lines. In
figure 5A we show as our mathematical model
where the parameters have been obtained through
the minimization of (3), is able to accurately de-
scribe the distribution of values of a 2D-GE spot.
Figure 5: A) In red is plotted the Gaussian distribution, in
green is plotted the spot non-saturated profile. B) In blue
is plotted the Gaussian distribution while in yellow is
plotted the spot saturated spot.
Finally, in figure 5B we show how the method
can effectively enable correct spots values recon-
struction fixing the spot saturation issue.
The procedure ran on more than 50 2D-GE im-
ages, each of them acquired at three different exposi-
tions (i.e. acquisition parameters).
3 CONCLUSIONS
Saturation of abundant spots is a general problem in
2-DE evaluation, in particular working with complex
samples like serum or plasma, which have a very
uneven protein distribution.
Currently available commercial software are not
able to perform 2D-GE image analysis in presence
of saturated spots. The only alternatives are to re-
move the saturated spots or rescan the gel image
with different acquisition parameters.
In this paper is presented a new approach for
treatment of over-saturated protein spots in 2D-GE.
Using experimental 2D-GE images we have demon-
strated that saturated protein spots can be found by
our algorithm.
Subsequently, we applied a Gaussian function to
calculate the real experimental spot volume (and
thus the correct protein expression) through the
reconstruction of intensity distribution of non-
saturated spots. The accuracy of reconstruction was
verified by comparing the same gel acquired with or
without saturated spots.
REFERENCES
Clark, B. N., Gutstein, H. B., 2008. The myth of auto-
mated, high-throughput two-dimensional gel analysis.
Proteomics, 8, 1197–1203
Daszykowski, M., Bierczynska-Krzysik, A., Silberring, J.,
Walczak, B., 2009. Avoiding spots detection in analy-
sis of electrophoretic gel images. Chemometrics and
Intelligent Laboratory Systems
Matthias Berth, M., Moser, F. M., Kolbe, M., Bernhardt,
J., 2007. The state of the art in the analysis of two-
dimensional gel electrophoresis images. Appl Micro-
biol Biotechnol 76:1223–1243
Maurer, M. H., 2007. Software analysis of two-
dimensional electrophoretic gels in proteomic experi-
ments. Current Bioinformatics, 2006, 1, 255-262
Miller, I., Crawford, J., Gianazza, E., 2006. Protein stains
for proteomic applications: Which, when, why? Pro-
teomics. 6, 5385–5408
Nonlinear Web Site, 2011. http://www.nonlinear.com/
support/progenesis/samespots/faq/saturation.aspx
Rashwan, S., Faheem, T., Sarhan, A., Youssef, B. A. B.,
2010. A Fuzzy-Watershed Based Algorithm for Pro-
tein Spot Detection in 2DGE images. IJCSNS Interna-
tional Journal of Computer Science and Network Se-
curity 254, VOL.10 No.5
Srinark, T., Kambhamettu, C., 2008. An image analysis
suite for spot detection and spot matching in two-
dimensional electrophoresis gels. Electrophoresis, 29,
706–715
Sternberg, S., 1983. Biomedical Image Processing, IEEE
Computer, January 1983.
Weingarten, P., Luter, P., 2005. Application of proteomics
and protein analysis for biomarker and target finding
for immunotherapy. Adoptive immunotherapy: meth-
ods and protocols. Humana Press
Wheelock, Å. M., Buckpitt, A. R., 2005. Software-induced
variance in two-dimensional gel electrophoresis image
analysis. Electrophoresis, 26, 4508–4520
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
338
