
3D CONFOCAL MICROSCOPY DATA ANALYSIS USING
LEVEL-SET SEGMENTATION WITH ALPHA DIVERGENCE
SIMILARITY MEASURE
Leila Meziou
1
, Aymeric Histace
1
, Fr´ed´eric Precioso
2
, Bogdan J. Matuszewski
3
and Franck Carreiras
4
1
ETIS UMR 8051 CNRS /ENSEA/ Cergy-Pontoise University, 95000 Cergy, France
2
I3S - UMR 6070 - CNRS/Nice Sophia Antipolis University, France
3
ADSIP Research Centre, University of Central Lancashire, Preston, U.K.
4
ERRMECe, Cergy-Pontoise University, 95000 Cergy, France
Keywords:
Image Segmentation, Active Contours, Alpha-divergence, Level-set, Confocal Microscopy.
Abstract:
Segmentation of cellular structures is of primary interest in cell imaging for a 3D reconstruction of cell shape.
Such an analysis provides crucial information about cell morphology and is instrumental in understanding
of biological processes leading to development of a particular pathology. The work presented in this paper
reports on a novel method for segmentation of cellular structures (nuclei and cell boundaries) from 3D single
channel actin tagged fluorescence confocal microscopy images. The proposed segmentation method uses
histogram-based image similarity measure in a level-set active-contour framework. The novelty of the method
is in application of the alpha-divergence distance measure which can be seen as a generalization of classic
Kullback-Leibler and χ
2
measures. The resulting alpha-divergence level-set formulation leads to a single front
evolution formula for both nuclei and cell boundaries segmentation, with no requirements for any enhancement
or preprocessing of acquired cell images (a monolayer of human cells (PNT2) culture).
1 INTRODUCTION
Segmentation of cellular structures is an essential tool
in cell imaging as it enables measurements which can
be used to track cell divisions or help to reconstruct
corresponding cell lineage tree providing data for cal-
culation of different parameters like cell proliferation
rate for instance. More specifically, the work pre-
sented in this paper has been carried out in a con-
text of analyzing changes of cell cytoskeleton prop-
erties in a response to ionizing radiation insult. The
final goal of this research effort is to better under-
stand cell bio-mechanical responses during cancer ra-
diation therapy. Indeed, actin tagged fluorescence
confocal microscopy imaging enables to character-
ize important properties of cytoskeleton, in particu-
lar actin filaments which are involved in many cellu-
lar processes like cell adhesion, locomotion, inter-cell
transport and general cell structural integrity, to name
a few. Nevertheless, due to a highly complex actin
appearance, a high level of noise and a strong non-
homogeneity of intensity and gradient information,
the segmentation of cell structures in such imaging
data, is a very challenging task. In this context, we
propose a (quasi-)automatic segmentation approach,
reducing to a minimum manual interventions – which
represent practical bottleneck when considering many
monolayer acquisition – in order to extract nuclei and
cell boundaries that provide spatial reference frame
for analyzing cytoskeleton changes.
To date, only few methods propose to address seg-
mentation of cell structures in fluorescence confocal
microscopy images (FCMI). In former approach pro-
posed in (Ortiz De Solorzano et al., 1999), authors fo-
cused on nuclei segmentations. In (Yan et al., 2008)
authors proposed cell segmentation in 2D-fluorescent
images with two channels (actin and nucleus tag-
ging) using a multiphase level-set combining Chan-
Vese (Chan and Vese, 2001) and geodesic active
contour models, together with repulsive force intro-
duced to prevent segmented cells from overlapping.
In (Mosaliganti et al., 2009; Zanella et al., 2010),
automated 3D cell segmentation from 3D confocal
acquisition of early Zebrafish embriogenesis is pro-
posed, two different fluorescent markers (red for nu-
clei and green for membrane) are used to easily dis-
405
Meziou L., Histace A., Precioso F., J. Matuszewski B. and Carreiras F..
3D CONFOCAL MICROSCOPY DATA ANALYSIS USING LEVEL-SET SEGMENTATION WITH ALPHA DIVERGENCE SIMILARITY MEASURE.
DOI: 10.5220/0003820004050409
In Proceedings of the International Conference on Computer Vision Theory and Applications (VISAPP-2012), pages 405-409
ISBN: 978-989-8565-03-7
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

criminate nuclei from cell membranes. In (Zanella
et al., 2010), authors introduced an adapted version
of the subjective surface technique (Sarti et al., 2002)
for surface reconstruction from missing boundary in-
formation whereas (Mosaliganti et al., 2009) use a
multiphase level-set based on propability correlation
functions.
Although our method is somewhat similar to (Yan
et al., 2008; Mosaliganti et al., 2009) focusing on
level-set framework, the objective is different since
our microscopic 2D images are extracted from a 3D
single channel confocal acquisition with only one flu-
orescent marker used for actin tagging making it a
more challenging problem. Nuclei and cell bound-
aries segmentations are considered as two separate
tasks but both use the same Partial Differential Equa-
tion (PDE) describing evolution of the active contour.
Finally, as it will be shown, no enhancement or pre-
processing will be required to segment acquired im-
ages.
The data used in this paper were obtained from
human prostate cells (PNT2). Once confluent cells
were fixed, actin were labelled with phalloidin-FITC
according to the manufacturers instructions (Invitro-
gen, UK). All imaging was carried out using a Zeiss
LSM510 confocal microscope. Fig. 1 shows some
images extracted at different slice levels from the
3D microconfocalacquisition of the monolayer PNT2
cell culture. The stack volume is defined on the
512×512×98grid of pixels each 0.21µm × 0.21 µm
× 0.11 µ in size.
The remaining of this paper is organized as fol-
lows: in Section 2, the alpha-divergence measure
and our histogram-based active contour segmentation
approach are explained together with the derivation
of the corresponding governing PDE in the level-set
framework; Section 3 focuses on experiments fol-
lowed by conclusions drawn in Section 4.
2 THEORETICAL FRAMEWORK
Originally proposed in (Kass et al., 1988), the basic
idea of the active contour is to iteratively evolve an
initial curve towards the boundaries of target objects
driven by the combination of internal forces, deter-
mined by the geometry of the evolving curve, and ex-
ternal forces, induced from the image. Image segmen-
tation methods using active contour are often derived
from a variational principle in which a functional de-
fined on contours encodes our knowledge about de-
sirable solution. The functional minimization leads to
a partial differential equation (PDE), constructed as
the Gateaux derivative gradient flow which steers the
evolution of the active contour.
In the particular framework of a region-based ac-
tive contour segmentation (Aubert et al., 2003) ,
several functional definitions have been proposed to
take into account the Probability Density Functions
(PDFs) of both the inner and outer regions of the
evolving curve. The corresponding variational crite-
rion is based on the minimization of a distance be-
tween PDFs calculated for the inner and outer regions
as defined by the evolving contour and predefined
reference PDFs of targeted and background objects.
Common distances used to compare PDFs are, for in-
stance, the χ
2
distance (Aubert et al., 2003) or the
Kullback-Leibler divergence (Lecellier et al., 2010).
In this paper, we propose an original histogram-based
active contour approach integrating alpha-divergence
as distance to minimize between two PDFs which can
be estimated parametrically or not. Let ˆq denotes an
estimated PDF and p a reference one for a particular
region Ω extracted from the image. As ˆq represent the
current PDF, it is non-parametrically estimated using
Parzen window method. This choice is motivated by
the fact that ˆq has to be differentiable for the next cal-
culation step of the minimization scheme. Distance
between ˆq and p using the alpha-divergences is now
defined as follows (Beirami et al., 2008):
D
α
( ˆqkp, Ω) =
Z
ℜ
n
ϕ
α
( ˆq(λ, Ω), p(λ))dλ , (1)
with ϕ
α
the cost function related to alpha-divergence
measure defined by:
ϕ
α
( ˆq(λ, Ω), p(λ)) =
1
α(1− α)
α ˆq(λ, Ω)+ (1− α)p(λ)
− [ ˆq(λ, Ω)]
α
[p(λ)]
1−α
, (2)
where α ∈ ℜ.
A complete study about the mathematical properties
of alpha-divergence can be found in (Beirami et al.,
2008). Nevertheless, considering Eq. (2), for spe-
cific values of α, some aforementioned standard dis-
tances can be connected to alpha-divergences, for in-
stance: D
2
(Ω) =
1
2
D
χ
2
(Ω), D
1
2
(Ω) = 2D
Hellinger
(Ω),
D
KL
(Ω) =
1
2
lim
α→0
D
α
(Ω) + lim
α→1
D
α
(Ω)
.
In the general framework of the histogram-based
active contours the alpha-divergence functional can
now be defined as follows for grayscale images (n =
1):
J
α
(Γ, Ω
in
, Ω
out
) = ξ
in
Z
ℜ
ϕ
α
( ˆq(λ, Ω
in
), p
in
(λ))dλ
+ ξ
out
Z
ℜ
ϕ
α
( ˆq(λ, Ω
out
), p
out
(λ))dλ
+ β
Z
Γ
ds (3)
VISAPP 2012 - International Conference on Computer Vision Theory and Applications
406

where Ω
in
and Ω
out
are respectively the foreground
(targeted object) and the background areas, Γ the
evolving boundary between Ω
in
and Ω
out
and ξ
in
, ξ
out
and β three positive weighting parameters. Consid-
ering now the standard level-set embedding function
U of J
α
, the Lagrangian minimization scheme of J
α
leads to the following associated PDE for active con-
tour evolution:
∂U
∂t
= δU
β∇·
∇U
|∇U|
−
ξ
in
|Ω
in
|
(A
in
−C
in
)
+
ξ
out
|Ω
out
|
(A
out
−C
out
)
, (4)
with
A
i
= ∂
1
ϕ
α
( ˆq(λ, Ω
i
), p
i
(λ)) ∗ g
σ
(I(x)), (5)
C
i
=
Z
ℜ
∂
1
ϕ
α
( ˆq(λ, Ω
i
), p
i
(λ)) ˆq(λ, Ω
i
)dλ,
where i = {in, out} and ∂
1
ϕ
α
denotes the first deriva-
tive order of ϕ
α
function with respect to ˆq, g
σ
is the
Gaussian kernel (with standard-deviation σ) used in
the Parzen window estimation of ˆq, I is the intensity
function of the segmented image at a pixel x. The
implementation of Eq. (4) is achieved with a semi-
implicit version of the Additive Operator Splitting
scheme.
3 EXPERIMENTAL RESULTS
In order to achieve the automatic segmentation of
both nuclei and cell boundaries, inner and outer refer-
ence PDFs, corresponding to the targeted structures,
are computed applying a standard three class (for
cell boundaries, cytoplasm and nucleus) Expectation-
Maximization (EM) algorithm on the equator slice
of the PNT2 acquisition. This rough classifica-
tion is good enough to define the reference PDFs
using Parzen window technique: the strategy is
then one class (“nucleus”, resp. “cell boundaries”)
against the two others (“cell boundaries+cytoplasm”,
resp.“nucleus+cytoplasm”). The segmentation pro-
cess is initialized on the equator slice (Fig. 1 mid col-
umn) through the monolayer. For both nuclei and cell
boundaries segmentations, the zero level-set initial-
ization of the function U
0
is given as a set of small
circles uniformly distributed across the whole slice.
Finally, the segmentation process is spread all along
the different slice level of the monolayer.
Fig. 1 summarizes results obtained for different
slices from the acquired 3D image stack and for dif-
ferent values of the parameter α. All tests are done
with the same optimal parameters ξ
i
(here ξ
in
= ξ
out
)
and β for both nuclei and cell boundaries (empirically
tuned on the equator slice image) in order to focus on
analysis the influence of the α parameter only.
Considering nuclei segmentation, as shown in
Fig. 1 in green, thanks to the level-set formulation
and to the integration of alpha-divergence measures,
all nuclei are detected during a single run of the algo-
rithm, even those with incomplete shape (at the bor-
der of image). This is a major advantage when com-
paring with the Chan-Vese approach which needs to
be carefully adapted (in its regularized form) to per-
form the same task as shown in some of our previ-
ous work (Meziou et al., 2011) on that topic. It is
also important to notice that the more accurate results
are obtained with non-standard values of α as shown
in Figs. 1. (b). where α = 0.75. The real difficulty
is to obtain a robust segmentation result of each nu-
cleus to avoid small structures segmentation within
cytoplasm reflecting the complexity of the cytoplasm-
nucleus boundary. For the higher values, false detec-
tions could appear and non significant small structures
are segmented as shown in Figs. 1. (e).
For each slice segmentation, an expert can man-
ually suppress false detected nuclei that can occur
when a hole representing an empty space between
cells is present in the cell culture for instance: Sta-
tistically speaking, the PDF of a hole is very close to
the PDF of a nucleus and can be difficult to differen-
tiate automatically.
In the case of cell boundaries, shown in red in
Fig. 1, segmentation is more demanding since even
for experts it is not always easy to visually identified
them: the non-homogeneity of the actin fluorescent
marker can strongly influence the pixel levels corre-
sponding to cell boundaries which could explain dif-
ficulties to get continuous contours. Results obtained
are similar to those obtained with Chan and Vese seg-
mentation which performed well for this particular
task and can be considered as a reference for that kind
of single channel acquisition. However, major advan-
tage of the proposed approach for cell boundaries seg-
mentation is the use of the same evolution criterion
(alpha-divergence measure) for both cell boundaries
and nucleus segmentations. Influence of the α param-
eter, as for nuclei segmentation, is interesting since
it can be noticed again that the most interesting re-
sults are obtained for a value of α that does not cor-
respond to standard histogram distance measures. In
terms of cell boundaries segmentation, the proposed
results have to be considered as prospective ones but
very encouraging. From an expert point of view, the
best results are obtained for a high value of α (for ex-
ample α = 1.5 shown in Figs. 1.(d)).
3D CONFOCAL MICROSCOPY DATA ANALYSIS USING LEVEL-SET SEGMENTATION WITH ALPHA
DIVERGENCE SIMILARITY MEASURE
407
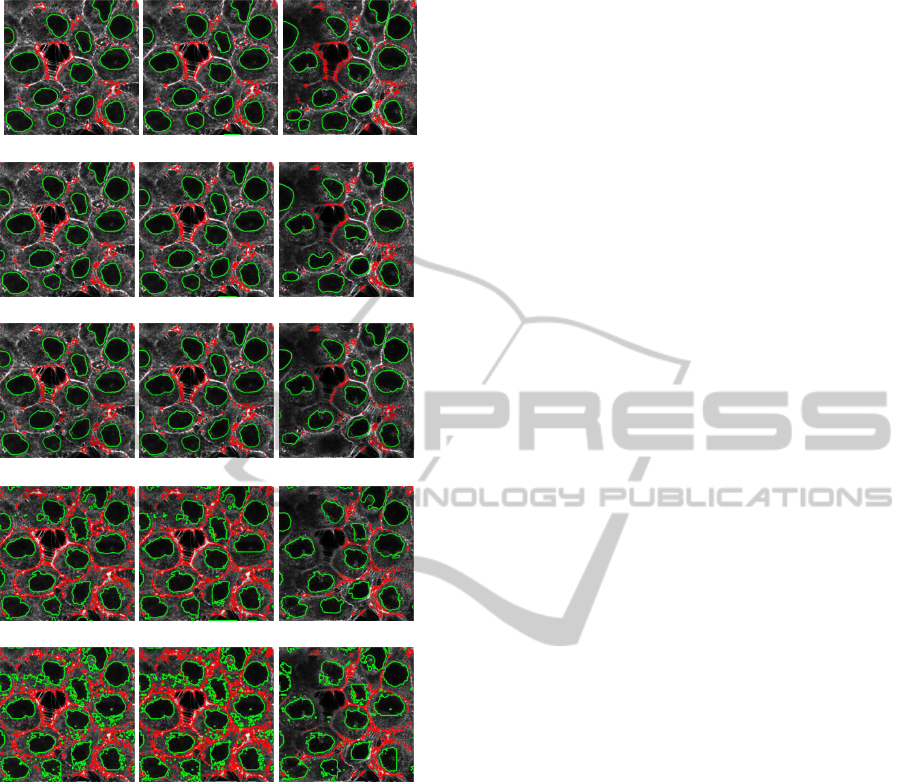
(a) α = 0.5.
(b) α = 0.75.
(c) Kullback-Leibler divergence
(d) α = 1.5.
(e) α = 2.
Figure 1: Segmentation of cell nuclei (in green) and cell
boundaries (in red) with different values of α (row) and for
different slice index in the acquired 3D image stack (col-
umn, from left to right slices from bottom, middle, and top
of the stack).
4 CONCLUSIONS
In this paper a novel histogram based level-set ac-
tive contours method is proposed for segmentation
of nuclei and cell boundaries in 2D images from a
3D single channel confocal microscopic acquisition
of a PNT2 cells monolayer. Introduction of alpha-
divergence measure within the variational framework
leads to a single evolution PDE for segmentation of
both nuclei and cell boundaries. The segmentation re-
sults obtained for the nuclei and cell boundaries, show
that the method enables to control in an efficient way
a range of distances that can be used in very different
segmentation scenarios. Moreover, compared to pre-
vious works, the proposed method does not require
any enhancement or preprocessing since the perfor-
mance of alpha-divergence measure can be adapted
to the level of the corrupting noise and if expert man-
ual interactions are permitted, they can be reduced to
a minimum. Additionally, as already mentioned, the
cell structure segmentation could be achieved in the
particular case where only actin is tagged in opposi-
tion to other recent studies reported in the literature.
It is envisaged that the future work will include: (i)
improvement of cell boundaries segmentation using a
local adaptation of α parameter based on noise char-
acteristics of each classes , and (ii) development of a
joint nuclei and cell boundaries 3D segmentation.
ACKNOWLEDGEMENTS
This work was supported by the UK Engineering and
Physical Sciences Research Council [TeRaFS project,
grant number EP/H024913/1].
REFERENCES
Aubert, G., Barlaud, M., Faugeras, O., and Jehan-Besson,
S. (2003). Image segmentation using active contours:
Calculus of variations or shape gradients? SIAM J.
Appl. Math., 63:2128–2154.
Beirami, A., Cevher, V., Bower, B., and Tsianos, K. (2008).
Proofs of alpha divergence properties. Technical Re-
port STAT 631 / ELEC 639, Rice University.
Chan, T. F. and Vese, L. A. (2001). Active contours without
edges. IEEE trans. on IP, 10(2):266–277.
Kass, M., Witkin, A., and Terzopoulos, D. (1988). Snakes:
Active contour models. Int. J. Comput. Vision,
V1(4):321–331.
Lecellier, F., Fadili, M., Jehan-Besson, S., Aubert, G.,
Revenu, M., and Saloux, E. (2010). Region-based ac-
tive contours with exponential family observations. J.
of Math. Imaging and Vision, 36(1):28–45.
Meziou, L., Histace, A., Precioso, F., Matuszewski, B.,
and Murphy, M. (2011). Confocal Microscopy Seg-
mentation Using Active Contour Based on Alpha-
Divergence. In Proceedings of ICIP 2011, pages
3138–3141.
Mosaliganti, K., Gelas, A., Gouaillard, A., Noche, R., Ob-
holzer, N., and Megason, S. (2009). Detection of spa-
tially correlated objects in 3d images using appear-
ance models and coupled active contours. In Proceed-
ings of MICCAI’09, pages 641–648, Berlin, Heidel-
berg. Springer-Verlag.
Ortiz De Solorzano, C., Garcia Rodriguez, E., Jones, A.,
Pinkel, D., Gray, J. W., Sudar, D., and Lockett, S. J.
(1999). Segmentation of confocal microscope images
VISAPP 2012 - International Conference on Computer Vision Theory and Applications
408

of cell nuclei in thick tissue sections. Journal of Mi-
croscopy, 193(3):212–226.
Sarti, A., Malladi, R., and Sethian, J. A. (2002). Subjective
surfaces: A geometric model for boundary comple-
tion. Int. J. Comput. Vision, 46(3):201–221.
Yan, P., Zhou, X., Shah, M., and Wong, S. T. C. (2008).
Automatic segmentation of high throughput rnai fluo-
rescent cellular images. IEEE Transactions on Infor-
mation Technology in Biomedicine, 12(1):109–117.
Zanella, C., Campana, M., Rizzi, B., Melani, C., San-
guinetti, G., Bourgine, P., Mikula, K., Peyri´eras, N.,
and Sarti, A. (2010). Cells segmentation from 3d con-
focal images of early zebrafish embryogenesis. IEEE
trans. on IP, 19(3):770–781.
3D CONFOCAL MICROSCOPY DATA ANALYSIS USING LEVEL-SET SEGMENTATION WITH ALPHA
DIVERGENCE SIMILARITY MEASURE
409
