
eBIOMICS
A Website Promoting Good Practices
and Specific Methods for using Bioinformatics Resources
Frédérique Lisacek
1
, Patrick Koks
2
, Pascale Berthault
3
, Grégoire Rossier
1
, Guy Bottu
4
,
Jacques van Helden
4
, Jean Sylvestre
3
, Jean-Pierre Kraehenbuhl
5
and Jack Leunissen
1
1
SIB Swiss Institute of Bioinformatics, Geneva, Switzerland
2
Wageningen University, Bioinformatics Laboratory, Wageningen, Netherlands
3
SDC Conseil & Edition, Paris, France
4
Université Libre de Bruxelles, Bruxelles, Belgium
5
Health Sciences eTraining Foundation (HSeT), Epalinges, Switzerland
Keywords: Bioinformatics, e-learning, -omics, Interactive Platform.
Abstract: eBiomics (www.ebiomics.org) is an e-learning interactive platform in bioinformatics intended as a support
to post-graduate students and scientists involved in -omics or systems biology projects. The heterogeneity
and the multitude of bioinformatics resources (databases or tools) make their individual use and
appreciation difficult though difficulties can be overcome through indexing examples of use in specific
contexts. eBiomics is presented as a didactic guide for an extensive range of on-line databases and tools
commonly referred to in -omics applications. In its self-training version, it was developed with a free
content management system and contains light textual information complemented with numerous
screenshots and visual information in a user-friendly environment. A collection of protocols and case
studies is indexed for finding representative and practical usages of each of the resources described. Most
references to on-line databases and tools are accessible in real time through cross-links. The content covers
main -omics related topics. Such a flexible tool allows keeping up with the constant evolution imposed by
frequent new releases of databases, upgrades of on-line software and regular changes of query interfaces,
which usually precludes from publishing helpful tutorials in books and manuals (otherwise rapidly
becoming obsolete). In addition, it offers access to a self-testing section. A complementing sister website
focused on biomedical academic studies includes part of the self-training content in a different environment.
1 INTRODUCTION
The increase in data generation techniques in the
Life Sciences (so called -omics techniques) makes it
possible to generate huge amounts of scientific data
in even a single experiment. As the cost of such
technology drops simultaneously, the generation of -
omics data comes within reach of many research
groups, exposing more scientists to these data. This
growth in data (techniques) is accompanied by a
subsequent increase in data resources and analysis
tools. The NAR bi-yearly special issues (database
(Galperin and Cochrane, 2011), web server (Benson,
2011)) have captured this trend in the last decade. It
is envisaged that researchers in the Life Sciences
now and in the future will no longer spend most of
their time in the laboratory (the data generation often
being outsourced to an external party), but behind a
computer to analyse the research obtained by an
experiment (Sboner, 2011).
All of this renders it virtually impossible for
students or established scientists that are unfamiliar
with these fields to gain a quick overview and insight
in which tools and/or databases are available and
which are most appropriate for a particular study or
technique. Furthermore, the speed of evolution of
technology renders large parts of practical textbooks
and manuals obsolete on a short-term basis. Teaching
and training requires regular reappraisal and
restructuring to keep up with latest developments.
This aspect is particularly true in bioinformatics as
exemplified in a recent issue of PLoS Computational
Biology partly devoted to bioinformatics education
(Via et al., 2011). The need for frequent update and
for reaching increasing numbers of learners has
logically supported the move to e-learning (Schneider
386
Lisacek F., Koks P., Berthault P., Rossier G., Bottu G., van Helden J., Sylvestre J., Kraehenbuhl J. and Leunissen J..
eBIOMICS - A Website Promoting Good Practices and Specific Methods for using Bioinformatics Resources.
DOI: 10.5220/0003893703860391
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (SSTB-2012), pages 386-391
ISBN: 978-989-8425-90-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

et al., 2011; Wright et al., 2011).
Virtual bioinformatics courses were initiated in
VSNS (de la Vega et al., 1996) or the EMBER
project (2001) mainly focused on sequence analysis.
The latter has been kept alive on and off since the
project end in 2005 highlighting the difficulty of
sustaining this type of effort. Even references
recognised as successful in a recent review (Wright
et al., 2011) like BioManager (Cattley et al., 2010) is
now discontinued. Consequently, a scheme for
knowledge sustainability is a central issue that needs
attending. Successful on-line teaching programmes
like those developed at the universities of
Manchester or Bielefeld (BiBiServ) cited in the
same review (Wright et al., 2011) tend to show that
rooting e-learning strategies into university courses
increases sustainability.
An early version of a self-learning website in
bioinformatics for proteomics (e-proxemis) was
developed in recent years by members of the SIB.
Despite positive feedback and a lifespan of several
years (a few thousand registrations during that time),
the platform was restricted to the proteomics domain
thereby remaining limited in reach (the genomics
and transcriptomics communities are much wider).
The expansion of –omics technology reinforced by
the frequent combination of more than one –omics
approach in biological studies impose a reflection on
the renewal of teaching strategies. This concern is
shared in OpenHelix (Williams et al, 2011) though
the tutorial format on this platform is not interactive.
The eBiomics project presented in this paper
strives to provide an e-learning environment in
which both students and experts can find
information relevant to their field of study. The
purpose of eBiomics is to familiarise users with
bioinformatics analysis flows in diverse -omics
applications. To that end, we revised the e-proxemis
pedagogical strategy to account for the spread of
automated pipelines and tied our effort to an existing
e-learning platform that is already used in master
programmes (provided by the HSeT (Health
Sciences eTraining) Foundation).
The resources that populate the eBiomics
catalogue were selected to reflect both the widely
recognised usage in a given –omics community and
the citations in our collection of case studies. In that
sense they are not bound to a set geographical origin
or to the respective services provided by the
institutes to which the authors belong. The catalogue
is not destined to become exhaustive in terms of
coverage of existing tools but to focus on a selection
that mirrors widely adopted and/or recommendable
practises.
Other content in eBiomics was originally prompted
by the following observation: most published work in
the field of –omics includes a compact description of
data analysis captured in a few paragraphs in the
Material and Methods section of the article. The
unfamiliar reader may recognise but also discover
software names or note references to well-known and
less known databases. The eBiomics platform
proposes the expansion of such condensed paragraphs
in detailed case studies to enhance the reading of an
article in two complementing ways. The approach is
known of “article-based learning”. On the one hand, it
provides a guide to learn about the most popular
individual resources as traditionally done in e-
learning platforms (EMBER, etc). On the other hand,
it illustrates their combined use in several contexts. In
fact, the latter approach assumes that data analysis
follows protocols in much the same way data
generation does. In eBiomics, protocols are presented
as flowcharts, text and put into the context of a
specific case study.
In short, eBiomics was built on past experience
from both EMBER, e-proxemis and on the expertise
of HSeT for grounding e-content in university
programmes. Two versions are hosted on two sites.
The full content is available for self-training
(currently at http://ebiomics.sdcinfo.com/) and
selected content for targeted article-based learning is
transferred to the HSeT site for academic use. The
present paper describes the full content and outlines
the basis of transfer to HSeT.
2 eBIOMICS PRINCIPLES
Besides creating an environment for e-learning and
specific use of bioinformatics resources, pedagogy
and long-term sustainability are the main focus of
eBiomics that constrained its design and
development. Even though a range of books
describes several aspects of bioinformatics (e.g.,
basics, algorithms), electronic media are more
adapted to assisting scientists in mastering day-to-
day usage of data analysis tools and reference
databases. Like books, an e-platform encourages
self-training; unlike books, e-content can easily and
rapidly be updated. Furthermore, the main advantage
of e-tools over books is interactivity, which is
essential for involving learners. Very few initiatives
were launched to take on the challenge.
2.1 Pedagogy for Self-training
The pedagogic strategy is two-fold. Firstly, it is
eBIOMICS - A Website Promoting Good Practices and Specific Methods for using Bioinformatics Resources
387

supported by guided navigation in the full content
website. The content of a page is centrally displayed
and the right side is populated with the variety of
related content available in the 5 sections of
eBiomics. Information content is thus accessible in
multiple ways. Secondly, it focuses on research
articles as the basis of self-training and e-learning
material for the HSeT website.
Figure 1 below illustrates the hierarchy of the
different sections in the full content website. The
structuring principle relies on zooming from the
global view of an analytical workflow to the basic
operation involved in a given step of analysis. The
top layer describes the many possibilities of building
a workflow given a set of analytical tasks. The next
layer describes one specific set of tasks and
associated tools which characterises an in silico
protocol. The next layer describes a case study
within which a protocol is used. A protocol can be
decomposed into steps each of which is viewed as
lower level single operation (specific if fully
described, generic if not).
Figure 1.
2.2 Content
For the first release, the mainstream bioinformatics
domains, namely genomics, proteomics,
transcriptomics and metabolomics, are covered.
Databases and tools can be categorised into one or
more of these domains or in the 'Sequence Analysis
and Annotation' “superdomain”. Indeed, sequence
analysis plays a central role in all –omics domains
and is therefore given a central place. As most of the
underlying data and techniques for analysing data
are also used in systems biology, we deem this
content as a valuable source for students and experts
in this research area as well.
For the full content site, eBiomics relies on the
flexibility of the content management system Drupal
to guarantee the ease of maintenance and update by
contributing authors. Update protocols keep the
maintenance task to a minimum when no major
change is associated with the new release of a
database or tool. They also help the rapid revision of
results that depend on database content (potentially
changing on a daily basis). When a new release
involves substantial changes, extra effort is
unavoidable but the repercussion of changes can be
controlled.
At this stage, the HSeT website only covers the
transcriptomics and proteomics domains with selected
content from its sister site, as explained below.
3 eBIOMICS IMPLEMENTATION
The description in the following 3.1 and 3.2 sections
focuses on the full content website designed for
interactive self-training. Section 3.3 outlines the
correspondence with the HSeT site.
3.1 Structure
The full content website is freely accessible from
www.ebiomics.org via registration for monitoring
purposes. To cater for users who reject the
obligation for registration, a login:demo
/password:demo was created.
As hinted in section 2.1, the website is composed
of several interconnected sections that can be
accessed through different interactive activities. The
5 main sections of eBiomics are:
1) Resources: this is a catalogue of selected
databases and data analysis software that are briefly
described and illustrated with examples. The
catalogue is ordered by categories mainly describing
the biological data (e.g., gene expression, proteins)
or the purpose of analytical approaches (e.g. pattern
search, structure prediction).
2) Conceptual Flowcharts: this is a collection of
clickable images picturing typical data analysis
flows in different -omics studies. No current large-
scale initiative produces datasets that are analysed
with one tool only. It is therefore important to stress
the various steps of analysis and show global views.
3) Protocols: this is a collection of recognised
approaches to problem-solving in common -omics
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
388

applications. Each step of an –omics protocol is
called a generic operation.
4) Case Studies: this is a collection of concrete
studies undertaken to address a biological question.
Since in the vast majority of cases, bioinformatics
analysis summarised in a published article is not
detailed, case studies expand and explain these short
summaries into self-sufficient accounts of
information found in selected publications. A case
study usually describes the implementation of a
specific protocol made of specific operations.
5) Exercises: self-evaluation space. Questions,
quizzes, problems and suggested solutions populate
this section.
3.2 Navigation Scheme
We now detail the various ways of browsing the
multiple categories of content.
3.2.1 Basics
a) To learn about the content of a database or the
purpose of a software tool only known by name, we
recommend looking for its description and examples
of use in the Resources section. Further context-
sensitive information can be accessed by clicking on
related content on the right panel of the page.
Example: the user has heard of Prosite but do
not really know what it is, then click on Resources,
find "Prosite" in the sub-section Databases, click on
"Prosite" and read content.
A resource is always described on the same
template answering the following questions:
- What is it?
- How and when to use?
- In a nutshell
b) To learn about the combined use of software
and/or databases or about the similarity of resources
we suggest checking the Conceptual flowcharts to
apprehend the chronology of tasks in the course of
analysis. Related content is available on the right
panel of the page.
Example: you use the Mascot software for
analysing mass spectrometry data and would like to
know of an alternative tool performing the same
analysis, then click on Conceptual flowcharts and
look at the yellow boxes featuring with the Mascot
box. Click on these boxes to read the content.
3.2.2 Advanced
a) To learn about the most typical data processing,
we encourage exploring the Protocols section.
Related content is available on the right panel of the
page.
b) To learn about the role of bioinformatics-based
analysis in concrete examples of research, we
recommend following the threads proposed in the
Case studies.
Example: you wish to understand the details of a
bioinformatics analysis as part of a global study
identifying disease biomarkers. These details are
given in a Case study where each important
processing step is singled out, explained and builds
up a complete story.
A case study is described on the same template
according to the following progression:
- Introduction
- Step 1 (specific operation 1)
- …
- Step n (specific operation n)
- Conclusion
A specific operation typically entails using one
piece of software or one type of database. It will
therefore be indicated as related content for the
description of that software/database in the Resource
section. A Case study is often based on
the
expansion of the few paragraphs summarising the
data analysis protocol in the Material and Methods
section of published articles describing large-scale
studies.
Finally, the Exercises section is designed to
evaluate the depth of a user's knowledge through
solving problems or answering quizzes. Each
resource or case study is related to relevant
exercises.
3.3 Article-based Learning
The HSeT website hosts its own version of eBiomics
and integrates part of the full content website in a
range of biomedical educative programmes. HSeT
provides tools for supervised e-training that are used
in several master programmes. HSeT mainly targets
medical science education and now relies on
eBiomics content for feeding the bioinformatics
section of its supervised e-training biomedical
programmes.
Practically, proteomics and transcriptomics Case
studies of the full content website were imported
into the HSeT website as simple HTML code along
with all related resource descriptions. Subsequently,
articles are annotated in the HSeT website to
enhance the medical/biological content with HSeT
tools. The final outcome is a multiply enriched
content.
eBIOMICS - A Website Promoting Good Practices and Specific Methods for using Bioinformatics Resources
389
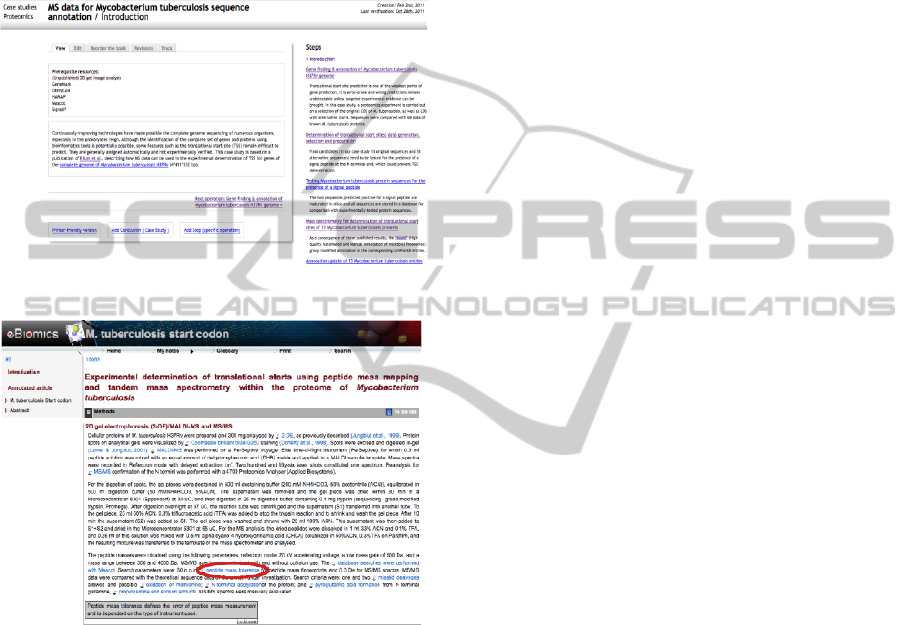
3.4 On-line Illustration
A live demo is the most convincing way of
illustrating the potential benefits of using eBiomics
and this will be detailed during the oral presentation.
As an introduction, the two screenshots below show
the full content and HSeT versions of the same case
study.
Figure 2: View in full content website.
Figure 3: View in HSeT website.
In the full content website (Figure 2), a case study
is introduced with a short text referencing the article
from which it is derived and navigation is detailed
on the right side of the page with a series of
clickable steps.
In the HSeT website (Figure 3), article-based
learning entails displaying the content by main
sections (here Methods) where terms are linked to
further information either in bioinformatics or in
medical science when appropriate. The example
shows that if “peptide mass tolerance” is clicked on
then the grey textbox below is displayed.
4 DISCUSSION
AND CONCLUSION
At present, the most common approach of untrained
scientists consists in running software with default
parameters and extracting data from databases upon
simple keyword search. In eBiomics, training is
regarded as the stepwise guidance of a trainee in
order to: (1) improve and refine his/her information
search strategies, (2) enhance his/her capacities for
critical assessment of results and (3) develop a sense
of extrapolation.
4.1 Target Users
eBiomics serves different user groups. New comers
to bioinformatics following a course in one of the –
omics domains present in eBiomics can answer
questions like: 'How are datasets analysed?', ‘what is
the purpose of this database?’, etc. Advanced users,
typically life science researchers, can answer
questions like: 'which database or tool is available
for my specific analysis or problem?', 'What are the
optimal settings for the algorithm I commonly use?'
or 'Can this or that method be applied in another
context?'
A next step in professionalising and increasing
the applicability of eBiomics to a wider range of
users can be the creation of a true Learning
Management System (LMS) behind the learning
environment. Plug-ins for this are readily available
and this makes Drupal a very suitable system for this
(Fitzgerald, 2009). By adding the possibility of
recording one's reading / learning progress,
bookmarking interesting or difficult sections and
storing test-results (for registered users and
teachers), the system will gain interest as a tool
matching needs for more regular teaching / training
/learning situations.
We also consider the option of expanding
content sharing beyond the existing partnership
potentially on the basis of solutions described in
(Romano et al, 2010).
4.2 Impact and Sustainability
Results in Life Sciences increasingly rely on
automated analysis of experimental data as -omics
sciences generate massive amounts of data. The
control of the quality of bioinformatics analysis has
become crucial for interpreting and valorising
results. In other words, those groups who will master
analysis tools best will produce the most interesting
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
390

results. In that respect, the expected impact of
eBiomics can be mostly evaluated in terms of
enhancing the quality of research. The reference to
standards and good practices is essential for students
and researchers involved in -omics applications.
The HSeT version of eBiomics contributes to
increasing its visibility and attracting a regular flow
of new comers. Further integration in master
programmes is considered in order to secure regular
use and longevity.
4.3 Prospects
The website was released with a critical mass of
content that should grow and improve in the coming
years as the partnership between the project partners
is maintained through multiple planned actions for
the popularisation of the website. Compliance with
the SCORM standard (legacy.adlnet.gov/
Technologies/scorm/) is nearly complete and will
also support easy transfer not only between the two
eBiomics sister websites but also potential other.
Further topics will be added as required by the
teaching programmes of partnering universities (in
Switzerland and the Netherlands so far). For
instance, specific systems biology tools like
stochastic modelling could be included in future
releases.
In conclusion, eBiomics is destined to provide an
environment that enables students and researchers in
the field of –omics and systems biology to be trained
or to train themselves, in order to make a better and
more efficient use of the available resources.
ACKNOWLEDGEMENTS
The development of eBiomics was supported by
grant #504187 of the EU Lifelong Learning
Programme Erasmus (multi-lateral project) and by
the Swiss Confederation. The maintenance and
expansion is supported by NBIC (Netherlands
BioInformartic Center).
REFERENCES
Attwood T et al., 2005. EMBER - A European Multimedia
Bioinformatics Educational Resource.
http://www.bioscience.heacademy.
ac.uk/journal/vol6/beej-6-4.aspx
Benson G., 2011. Nucleic Acids Research annual web
server issue in 2011. Nucleic Acids Research, 39,
p.W1-W2
Cattley S, Arthur JW, 2007. BioManager: the use of a
bioinformatics web application as a teaching tool in
undergraduate bioinformatics training. Brief
Bioinform., 8(6):457-65
de la Vega, F. M., Giegerich, R. & Fuellen, G., 1996.
Distance education through the Internet: the GNA-
VSNS biocomputing course. Pacific Symposium on
Biocomputing, pp.203-215.
e-proxemis, Unpublished http://e-proxemis.expasy.org/
Fitzgerald, B., 2009, Drupal for Education and E-learning,
Teaching and Learning in the Classroom using Drupal
CMS, Packt Publishing, Birmingham, UK.
Galperin, M. Y. & Cochrane, G. R., 2011. The 2011
Nucleic Acids Research Database Issue and the online
Molecular Biology Database Collection. Nucleic Acids
Research 39, p.D1-D6.
Romano, P., Giugno, R. & Pulvirenti, A., 2011. Tools and
collaborative environments for bioinformatics
research, Brief. Bioinf. 12(6):549 -561
Sboner et al., 2011. The real cost of sequencing: higher
than you think! Genome Biology, 12(8):125
Schneider, M.V. et al., 2010, Bioinformatics training: a
review of challenges, actions and support
requirements. Brief. Bioinform. 11(6):544-51
Via, A. et al., 2011. Ten Simple Rules for Developing a
Short Bioinformatics Training Course. PLoS Comput
Biol, 7(10), p.e1002245
Williams JM et al., 2010. Openhelix: bioinformatics
education outside of a different box. Brief.
Bioinform. 11(6):598-609
Wright VA et al., 2010, Bioinformatics training: selecting
a relevant content management system – an example
from the EBI. Brief. Bioinform. 11(6):552-62
eBIOMICS - A Website Promoting Good Practices and Specific Methods for using Bioinformatics Resources
391
