
TASK SCHEDULING IN A FEDERATED CLOUD
INFRASTRUCTURE FOR BIOINFORMATICS APPLICATIONS
C. A. L. Borges, H. V. Saldanha, E. Ribeiro, M. T. Holanda, A. P. F. Araujo and M. E. M. T. Walter
Department of Computer Science, University of Brasilia, Brasilia, Brazil
Keywords:
Federated Cloud Computing, Task Scheduling, Resource Allocation, Bioinformatics.
Abstract:
Task scheduling is difficult in federated cloud environments, since there are many cloud providers with distinct
capabilities that should be addressed. In bioinformatics, many tools and databases requiring large resources
for processing and storing enourmous amounts of data are provided by physically separate institutions. This
article treats the problem of task scheduling in BioNimbus, a federated cloud infrastructure for bioinformatics
applications. We propose a scheduling algorithm based on the Analytic Hierarchy Process (AHP) to perform
an efficient distribution for finding the best resources to execute each required task. We developed experiments
with real biological data executing on BioNimbus, formed by three cloud providers executing in Amazon
EC2. The obtained results show that DynamicAHP makes a significant improvement in the makespan time of
bioinformatics applications executing in BioNimbus, when compared to the Round Robin algorithm.
1 INTRODUCTION
Cloud computing is a resource delivery and usage
model to get resources (hardware, software, appli-
cations) through on-demand and at scale network
as services in a shared, multi-tenant and elastic en-
vironment (Li and Guo, 2010; Buyya and et al.,
2009). Computational capabilities in clouds rely
in the concept of virtualization, which is based on
several virtual machines providing services to con-
sumers. Cloud computing aims to offer virtually un-
restricted pay-per-use computational resources and no
needs to manage the underlying infrastructure. Re-
cently, the model of federated cloud (inter-cloud or
cross-cloud) was proposed, being generally defined as
a set of cloud providers, public and private, connected
through the Internet (Bernstein and et al., 2009; Ce-
lesti and et al., 2010; Marshall et al., 2010).
On the other hand, high performance genome se-
quencing technologies produce large amounts of bi-
ological data in hundreds of genome projects around
the world. A single genome project generates giga-
bytes of data, which have to be processed by large
and time consuming programs that also generate more
gigabytes of data, so requiring terabytes of storage
to store all these biological information. Genome
projects and related bioinformatics analyses are sup-
ported by workflows composed of tools and databases
that are usually stored in physically separate centers.
In this context, we previously proposed BioNim-
bus (Saldanha and et al., 2011), a cloud architecture
for bioinformatics applications, capable of transpar-
ently creating a computing environment that supports
dynamic expansion of resources (data storage, ser-
vices, virtual machines, CPUs) so that it can handle
variations in the service demands frequently found in
genome projects or in bioinformatics analyses.
Optimal use of computational resources should be
done according to a user demand, which is particu-
larly difficult in federated clouds, since resources of
distinct clouds may frequently be included to or re-
moved from the environment. Then, task schedul-
ing in federated clouds is a big challenge, since many
environment variables should be considered to guar-
antee efficient scheduling. Besides, the problem of
task scheduling on heterogeneous systems is NP-hard.
In this work, we study the problem of task schedul-
ing in federated clouds. The objective is to pro-
pose the dynamic resource allocation algorithm Dy-
namicAHP, based on the Analytic Hierarchy Process
(AHP) (Saaty, 1990), including it in BioNimbus.
This paper is organized as follows. Section 2 de-
scribes basic concepts of bioinformatics and federated
cloud computing, and presents BioNimbus. Section 3
proposes DynamicAHP. Following, Section 4 shows
how DynamicAHP was implemented in BioNimbus,
describes the experiments, presents the results, and
finally discusses related work. In Section 5, we con-
clude and suggest future work.
114
A. L. Borges C., V. Saldanha H., Ribeiro E., T. Holanda M., P. F. Araujo A. and E. M. T. Walter M..
TASK SCHEDULING IN A FEDERATED CLOUD INFRASTRUCTURE FOR BIOINFORMATICS APPLICATIONS.
DOI: 10.5220/0003932801140120
In Proceedings of the 2nd International Conference on Cloud Computing and Services Science (CLOSER-2012), pages 114-120
ISBN: 978-989-8565-05-1
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

2 BACKGROUND
Sequencing a DNA is the task of obtaining the bases
(A, C, G and T) of the so-called short-read sequences
(SRS), which are fragments belonging to the chromo-
somes of one or more organisms in a genome project.
DNA sequencing was strongly improved by the new
technologies developed by automatic sequencers like
Illumina and 454 Roche, among others. These se-
quencers are capable to produce millions of SRS of
one or more genomes in only one sequencing round,
each one of the SRS with lengths from 30 to 1,000
bases, depending on the adopted technology.
To evaluate the volume of data to be analyzed in a
genome project, we present two examples. Filichkin
et al. (Filichkin and et al., 2010) worked with approxi-
mately 271 millions SRS sequenced by Illumina, each
fragment with 32 bases. These bases were mapped to
a relatively short genome of the Arabidopsis thaliana
plant (≈ 120 millions of bases), with the objective
of identifying alternative splicing. Other research
groups (Pan and et al., 2008; Sultan and et al., 2008)
had the objective of identifying alternative splicing of
about 15 millions SRS, mapping them to the human
genome of approximately 3 billions of bases.
In this scenario, many projects were developed
for using cloud computing in bioinformatics aplica-
tions (Schatz, 2009; Langmead et al., 2010; Wall and
et al., 2010; Angiuoli and et al., 2011; Pratt and et al.,
2011; Zhang and et al., 2011). However, working
with a single cloud environment can be restrictive,
mostly due to execution in private companies, absent
of mechanisms to treat failures, and the fact that cur-
rent clouds are not flexible enough to allow unantici-
pated usages, which usually appear in bioinformatics.
Federation is a particular research area in cloud
computing. Celesti et al. (2010) note that middle-
ware implementations (OpenQRM, 2011; Bresnahan
and et al., 2011; Nurmi and et al., 2009) lack fed-
eration features, and explore general concepts about
cross-cloud federation. Cloud computing virtualiza-
tion allows increasing of computational resources and
reducing IT service costs by hiding the underlying in-
frastructure with a logical layer between the physi-
cal infrastructure and the computational processes. In
the cross-cloud federation, each cloud provider can
enlarge its virtualization resources demanding further
computing and storage capabilities to other clouds
transparent to users. Celesti et al. also point that the
implementation of a cross-cloud federation is not triv-
ial, although its clear advantages, since clouds are het-
erogeneous and dynamic, federation models are de-
signed for static environments and agreements among
the partners are required to create the federation.
In a previous work, we proposed BioNimbus (Sal-
danha and et al., 2011), a cloud computing infrastruc-
ture for managing bioinformatics tasks, designed to
be flexible and fault tolerant. The objective was to of-
fer the illusion that computational resources would be
unrestricted or, in other words, computational or stor-
age space demands would be always provided to the
users. In order to reach these objectives, we improved
BioNimbus using the federated cloud model (Fig-
ure 1). The infrastructure allows to integrate phys-
ically separate platforms, each modelled as a cloud,
which means that independent, heterogenous, pri-
vate/public clouds providing bionformatics applica-
tions could be integrated into a single federated cloud.
In BioNimbus, the resources of each user can be max-
imally used, but if more are required, other clouds can
be requested to participate, in a transparent way, so
that BioNimbus virtual resources amount are enlarged
by computational and data storage capabilities of all
the clouds forming the federation. A plug-in maps the
communication between a cloud provider integrating
the federation and the management services, so pro-
viding a simple and efficient way to include a new
cloud provider in BioNimbus.
Management services are implemented in the
BioNimbus core, which offers computational re-
sources such as virtual machines, data storage and
networks. A web interface was created to facilitate
the communication with users. Details of the BioN-
imbus core main services are:
• Discovery Service: identifies service providers
and consolidates information about storage ca-
pacity, processing, network latency and resource
availability;
• Monitoring Service: verifies if a requested service
is available in a cloud provider, searching for an-
other cloud in the federation if it is not; receives
the tasks to be executed from the job controller,
and sends them to the scheduling service to be
distributed, guaranteeing that all the tasks of a
process are really executed; informs the job con-
troller when a task successfully executes;
• Storage Service: coordinates the storage strategy
of the files consumed and produced by the exe-
cuted tasks, deciding about distribution, replica-
tion and file access control among the services;
• Security Service: guarantees integrity among the
distinct tasks executed in the federated clouds;
• Fault Tolerance Service: guarantees that all the
core services are always available. For this, mes-
sages for all the services are sent, and if some of
them do not react, initiates an election algorithm
to execute the service again in another machine;
TASKSCHEDULINGINAFEDERATEDCLOUDINFRASTRUCTUREFORBIOINFORMATICSAPPLICATIONS
115

Core
Monitoring
Service
Scheduling
Service
Storage
Service
Fault Tolerance
Service
Security
Service
Discovery
Service
Plug-in 1
Communication Interface (P2P)
MapReduce
Cloud
Providers
Plug-in 2
GridEngine
Plug-in n
Amazon EC2
Interpro
Bowtie
Bowtie
BLAST
Application
GUI – Graphical User Interface
Workflow Filtering Assembly Mapping Annotation
TopHat
Interpro
...
Figure 1: The BioNimbus federated cloud architecture.
• Scheduling Service: dynamically distributes tasks
among cloud providers. For this, it maintains
the register for the allocated tasks, controls each
cloud provider load, and redistributes the tasks
when the resources are overloaded.
Heterogeneity of the cloud providers is another
important aspect, which means that resources can
vary according to available services, storage capac-
ity and processing speed. Then, an efficient scheduler
plays a fundamental role in the performance of appli-
cations executing in clouds (Ergu and et al., 2011).
This is the focus of this work, the proposal of an effi-
cient dynamic task scheduler for federated clouds.
3 DynamicAHP
3.1 Analytic Hierarchycal Process
The Analytic Hierarchycal Process (AHP)
method (Saaty, 1990) aims at helping humans
to make decisions. Our algorithm is based on AHP,
since a decision making process, in many aspects,
resembles a scheduling algorithm. Saaty’s method
consideres human subjective factors, presenting
a fundamental scale to rank the importance of a
particular factor relative to another. This ranking
generates a matrix (Table 1) containing comparisons
among the factors.
Table 2 shows an example of how to use the funda-
mental scale matrix to create a comparison matrix. In
line A, the reciprocal values of object A relative to A,
Table 1: The fundamental scale proposed by Saaty (1990).
Intensity of Definition
importance
1 Equal importance
3 Weak importance of one
related to another
5 Essential or strong importance
7 Demonstrated importance
9 Absolute importance
2,4,6,8 Intermediate values between
the two adjacent judgments
Reciprocals If activity i has one of the above
of above non zero nonzero numbers assigned to it,
when compared to activity j,
then j has the reciprocal value
when compared to i.
B and C are 1, 3 and 2, respectively. Analogously, in
column A, the reciprocal values of object A relative to
A, B and C are 1, 1/3 and 1/2, respectively. Priority
column is computed as the sum of all the values of a
line divided by the sum of all the values of the matrix.
Then, Priority(A) = (1 + 3 + 2)/(1 + 3 + 2 + 1/3 +
1 + 1 + 1/2 + 2 + 1) = 0.507.
Table 2: A comparison matrix example.
A B C Priority
A 1 3 2 0.507
B 1/3 1 1 0.197
C 1/2 2 1 0.296
3.2 DynamicAHP Scheduler
DynamicAHP algorithm aims to minimize the
CLOSER2012-2ndInternationalConferenceonCloudComputingandServicesScience
116

makespan of tasks running on a federated cloud plat-
form, and was designed to reach the following charac-
teristics: makes scheduling decisions at runtime so it
is dynamic; performs task scheduling without having
any prior knowledge about task characteristics; and
does load balancing even when new cloud providers
are integrated in the federation.
Our scheduler creates metrics based on Saaty’s
(1990) fundamental scale, but instead of limiting the
fundamental scale to natural numbers from 1 to 9 and
their inverses, we will consider real numbers from 0 to
∞ (represented in a computer by the maximum value
of a float number). This scale states that a value v
much greater than a value w means that v presents a
higher priority when compared to w. In our algorithm,
the scheduling choice is based on ordering resources
considering their processing power, latency and re-
source reliability. These parameters were used to sim-
ulate the effect of executing a task as fast as possible
associated to a cost that should be as low as possible.
This strategy infers a “cost” to execute a task consid-
ering its size and the available resources provided by
each cloud provider. After evaluating these “costs”,
the tasks are descendingly sorted by their input sizes,
and the largest task is attributed to the best resource,
and so forth, until all tasks are scheduled.
In BioNimbus, the resources are highly unpre-
dictable. The computers are heterogenous, and the
workload is subject to constant changes. Besides, the
resources can be available or not at any moment, as
they belong to different cloud providers. The only
known information is the number of processors (vir-
tualized or not), the latency of the network, the maxi-
mum available disk size and the uptime (time that the
service is available in BioNimbus). BioNimbus can
execute many different bioinformatics applications,
such as Bowtie (Langmead and et al., 2009), a tool
that maps SRS to a reference genome.
In this scenario, the DynamicAHP scheduler
should first consider if the service requested by a task
is available, seeking a list of available services of the
discovery service. If the requested service is in this
available service list, the corresponding task will be
included in a scheduling list specific for the requested
service. If it is not, this task can not be executed,
and it is removed from the list, not being considered
for the rest of the decision making. All these tasks
will be kept in a pending job list until one of the fed-
erated clouds provides the required application, or a
new cloud provider is integrated in the federation. So
far, each service has a list of available tasks and re-
sources.
The next step is to indicate the priority of each
task and the importance of each resource. The task
is measured by the size of its executable file. The
process of defining the importance of each resource is
given by three factors: latency, uptime, total number
of available processors.
The DynamicAHP scheduler implemented in
BioNimbus analyzes the tasks to be executed, dynam-
ically allocates resources to execute them, and sends
a message to the monitoring service. The monitoring
service sends the job request to the plug-in service
and verifies the state of each task in each resource,
keeping the scheduler service informed.
The previously mentioned three factors (latency,
uptime, number of available processor) are numer-
ically represented. Each factor receives a relative
value, obtained considering a Saaty’s adapted funda-
mental scale. In details, DynamicAHP compares each
pair V
a
and V
b
, composed of factors and resources,
and computes the value for the fundamental scale as:
1 V
a
= 0 and V
b
= 0,
MAX FLOAT V
b
= 0,
V
a
V
b
otherwise.
For example, consider three resources, A, B and C,
with three factors (latency, uptime, quantity of avail-
able processors) influencing the total execution time
(Table 3). These factors should be compared to de-
cide which one is the best resource.
Table 3: Values associated to resource factors.
Resource Latency Uptime Available/Busy/
# Processors
A 13.0 0 12/0/0.084
B 10.0 0 4/0/0.25
C 14.0 0 4/0/0.25
Tables 4 and 5 show the comparisons of latency
and number of processors. The uptime factor will
not be considered since its value is 0 for a cloud
provider entering in the federation. To avoid division
by zero when there are no busy processors, and tak-
ing resA.BP as the number of busy processors (BP)
and resA.T P as the total number of processors (TP)
of resource A (resA), and analogously for resource B,
the number of available processors are computed as:
A ←
1.0
resA.T P
and B ←
1.0
resB.T P
(resA.BP = 0) and
(resB.BP = 0),
A ←
resA.BP
resA.T P
and B ←
resB.BP
resB.T P
otherwise.
Lower latency reduces the data transmission time,
while a greater number of available processors indi-
cates higher available processing capacity. So, the
previous formulas can be used for latency. Similarly,
for the uptime factor, a higher value is advantageous,
TASKSCHEDULINGINAFEDERATEDCLOUDINFRASTRUCTUREFORBIOINFORMATICSAPPLICATIONS
117

Table 4: Comparison matrix for latency.
A B C Priority
A 1.000 0.770 1.076 0.310
B 1.300 1.000 1.400 0.403
C 0.928 0.714 1.000 0.287
Table 5: Comparison matrix for number of processors.
A B C Priority
A 1.000 3.000 3.000 0.600
B 0.334 1.000 1.000 0.200
C 0.334 1.000 1.000 0.200
since it indicates that the service is available for a long
time and seems to be more reliable.
Table 6 shows global values, each one computed
by multiplying each factor resource priority (shown
in Tables 4 and 5), and indicates the priority ordering
as A > B > C. Despite resource A is not the best in
terms of latency, it is much better in the number of
available processors. Both B and C have the same
number of available processors, but resource B has a
better latency. If global priority is equal for more than
one resource, a mechanism similar to Round Robin
algorithm is activated to choose less used resources.
Table 6: Priority global values.
Latency CPUs Total
A 0.310 0.600 0.186
B 0.403 0.200 0.080
C 0.287 0.200 0.057
4 DISCUSSION
4.1 Implementation Details
BioNimbus and the DynamicAHP scheduler were
both implemented in Java. As stated earlier, BioN-
imbus architecture is divided into services. To exe-
cute a job, BioNimbus sends a request that activates
the scheduling service, which schedules and executes
the job. The scheduling service is implemented as
a thread, which runs in intervals of 15 seconds and
updates the status of all the cloud resources (cloud
map). This service is activated by a new job request.
Each job is inserted in a pending jobs list used by
the scheduling policy to choose the cloud where the
job will be executed. The scheduling policy uses the
cloud map to get information like latency, uptime,
number of busy cores, number of total cores or any
other resource information. The policy is defined in a
configuration file. A schedule method receives a list
containing each job’s information, like input and out-
put files, services to run or other job information, and
chooses the cloud where this particular job will be
executed. In this work, the implemented scheduling
policies’s were DynamicAHP and Round Robin.
4.2 Experiments
4.2.1 Cloud Providers, Tools and Data
To simulate three cloud providers, Hadoop infrastruc-
tures, with 12, 8 and 4 processors each, were created
at the Amazon EC2 (LLC, 2011). These infrastruc-
tures had about 2.4 terabytes of storage, implemented
through the Apache Hadoop Distributed File System
(HDFS). BioNimbus plug-ins executed on a virtual
machine to control each Hadoop infrastructure. Be-
sides, BioNimbus management services executed on
the largest infrastructure (12 processors).
The experiments consisted in executing Bowtie.
We note that it is straightforward to include other
applications, since our algorithm does not consider
any characteristic particularly related to an applica-
tion available in the cloud. In BioNimbus, Bowtie
was available as a service on each infrastructure
node (cloud provider) through the plug-ins. SRS of
the NCBI Mycobacterium tuberculosis genome were
mapped to the same organism’s genome. SRS were
distributed in three different files, with approximately
100, 200 and 300 MB, which were used as input files
to the experiments. For each one of these three input
files, groups of 20 jobs were created to simulate jobs
with different sizes. These jobs were sent to BioN-
imbus in groups of 5 jobs, in intervals of 60 seconds
between successive groups of jobs. This time interval
was used to simulate requests normally sent by differ-
ent users. The first job had an input of 300 MB, the
second 200 MB, the third 100 MB, and so forth.
4.2.2 Results
DynamicAHP presented a better makespan when
compared to Round Robin scheduler, as shown in Fig-
ure 2. Makespan was measured as the time the user
started the job until the time the job is finished. Fig-
ure 3 shows that DynamicAHP lowered the schedul-
ing time to execute the jobs, in almost all the cases,
when comparing to RoundRobin.
Round Robin overloaded the 4x cloud provider,
while DynamicAHP overloaded the 12x cloud
provider. Round Robin distributes its jobs evenly, so
that the 4x cloud provider becomes overloaded, al-
though the 12x cloud provider is capable of running
more jobs and finishes faster. In contrast, Dynam-
icAHP can check the resource status, and then can
CLOSER2012-2ndInternationalConferenceonCloudComputingandServicesScience
118
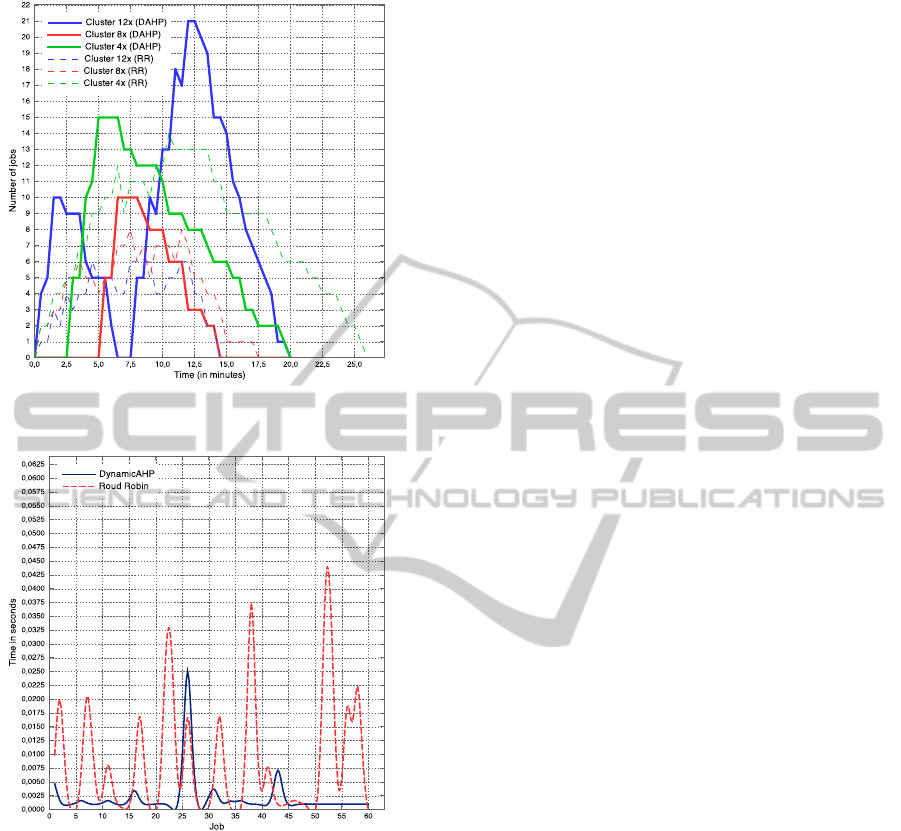
Figure 2: DynamicAHP and RoundRobin makespan and
load comparison.
Figure 3: Schedule time of DynamicAHP and RoundRobin.
run the job in the cloud provider with more available
processors, sending less jobs to the 4x cloud provider
and more jobs to the 12x cloud provider. This way,
DynamicAHP led to a more efficient job execution,
having executed all the 60 jobs in 19min36sec, while
Round Robin executed in 25min37sec.
Tasks are distributed by DynamicAHP in pre-
defined intervals, which includes new submitted
tasks. Besides, if new cloud providers enter in the
federation, integrated by BioNimbus, the scheduler
immediately consider them as new resources.
4.3 Related Work
Scheduling on cloud platforms has been extensively
studied. The well known Hadoop scheduler dis-
tributes scheduled tasks sequentially using FIFO. To
achieve data locality, for each idle server, the sched-
uler makes a greedy search for a data-local task in
the head-of-line job, allocating it to the server. This
work has one major drawback since it does not con-
sider resource heterogeneity, while the DynamicAHP
scheduler takes into account factors such as latency,
uptime and number of processors.
Henzinger et al. proposed a environment that ex-
ploits scalable static scheduling techniques to provide
a flexible pricing model, i.e., a tradeoff among dif-
ferent degrees of execution speed and price at the
same time. The authors noted that large scheduling
latencies make static scheduling impractical for large
clouds (Henzinger and et al., 2011). In this work, we
discuss a dynamic scheduling algorithm.
Some scheduling algorithms are based on stochas-
tic integer programming. Li and Guo (2010) pro-
posed an optimization model based on stochastic
integer programming to address the SLA (Service
Level Agreement) aware resource composition prob-
lem. Chaisiri et al. applied stochastic integer pro-
gramming for a resource provision optimization prob-
lem (Chaisiri et al., 2009). Their algorithm minimizes
the total cost of resource provision in a cloud envi-
ronment. However, they do not consider the notion
of SLA, which is one of the most important business
notion in cloud computing. Both scheduling methods
consider a single cloud platform, while our scheduler
considers federated cloud plataform.
Mehdi et al. devised an algorithm for finding a
fast mapping using genetic algorithms with an “ex-
ist if satisfy” condition to speed up the mapping pro-
cess and to ensure that all the task deadlines are re-
garded (Mehdi and et al., 2011). In contrast, Dynam-
icAHP makes decisions considering each task, and
continuously monitores the federated cloud environ-
ment for future decisions.
5 CONCLUSIONS
In this work, we proposed DynamicAHP, a task
scheduling algorithm to federated clouds, using sim-
ple parameters simulating “costs” (number of proces-
sors, latency and uptime). We realized experiments
with real biological data in BioNimbus, a federated
cloud architecture to execute bioinformatics applica-
tions. Results showed that DynamicAHP made a sig-
nificant improvement in makespan time without in-
creasing overload, when compared to Round Robin.
We plan to investigate how to include in Dynam-
icAHP more sophisticated methods to predict which
cloud provider would be better to execute a specific
TASKSCHEDULINGINAFEDERATEDCLOUDINFRASTRUCTUREFORBIOINFORMATICSAPPLICATIONS
119

task using a machine learning approach. Parameters
based on SLA associated to each task can also im-
prove our algorithm. A “cost” could be included as
one of the scheduling resources in DynamicAHP, as
well as weights for the factors (latency, uptime, avail-
able processors) that would dynamically change to
adapt to the cloud conditions. It is simple to include
in BioNimbus more cloud providers that will be au-
tomatically considered by our scheduler, and prelimi-
nary scalability analysis showed that our algorithm is
robust, but more conclusive tests should be done.
REFERENCES
Angiuoli, S. V. and et al. (2011). Resources and costs
for microbial sequence analysis evaluated using vir-
tual machines and cloud computing. PLoS ONE,
6(10):e26624.
Bernstein, D. and et al. (2009). Blueprint for the intercloud
- protocols and formats for cloud computing interop-
erability. Internet and Web Applications and Services,
International Conference on, 0:328–336.
Bresnahan, J. and et al. (2011). Cumulus: An open source
storage cloud for science. In 2nd Workshop on Scien-
tific Cloud Computing - ScienceCloud 2011, San Jose,
CA, United States. http://www.nimbusproject.org/.
Buyya, R. and et al. (2009). Cloud computing and emerg-
ing it platforms: Vision, hype, and reality for deliv-
ering computing as the 5th utility. Future Generation
Computer Systems, 25(6):599–616.
Celesti, A. and et al. (2010). How to enhance cloud ar-
chitectures to enable cross-federation. In IEEE 3rd
International Conference on Cloud Computing, pages
337–345. IEEE Computer Society.
Chaisiri, S., Lee, B.-S., and Niyato, D. (2009). Opti-
mal virtual machine placement across multiple cloud
providers. In APSCC 2009, Services Computing Con-
ference, pages 103–110. IEEE.
Ergu, D. and et al. (2011). The analytic hierarchy process:
task scheduling and resource allocation in cloud com-
puting environment. The Journal of Supercomputing,
pages 1–14. doi:10.1007/s11227-011-0625-1.
Filichkin, S. A. and et al. (2010). Genome-wide mapping of
alternative splicing in Arabidopsis thaliana. Genome
Research, 20(1):45–58.
Henzinger, T. A. and et al. (2011). Static scheduling in
clouds. In HotCloud 2011. USENIX Association.
Langmead, B. and et al. (2009). Ultrafast and memory-
efficient alignment of short DNA sequences to the hu-
man genome. Genome Biology, 10(3):R25+.
Langmead, B., Hansen, K., and Leek, J. (2010). Cloud-
scale RNA-sequencing differential expression analy-
sis with Myrna. Genome Biology, 11(8):R83.
Li, Q. and Guo, Y. (2010). Optimization of resource
scheduling in cloud computing. In SYNASC’2010,
12th International Symposium on Symbolic and Nu-
meric Algorithms for Scientific Computing, pages
315–320. IEEE Computer Society.
LLC (2011). Amazon Elastic Compute Cloud EC2.
http://aws.amazon.com/ec2/. Accessed Nov 22, 2011.
Marshall, P., Keahey, K., and Freeman, T. (2010). Elastic
site: Using clouds to elastically extend site resources.
Cluster Computing and the Grid, IEEE International
Symposium on, 0:43–52.
Mehdi, N. A. and et al. (2011). Impatient task mapping
in elastic cloud using genetic algorithm. Journal of
Computer Science, 7:877–883.
Nurmi, D. and et al. (2009). The eucalyptus open-source
cloud-computing system. In 9th IEEE/ACM Interna-
tional Symposium on Cluster Computing and the Grid,
CCGRID’09, pages 124–131, Washington, DC, USA.
IEEE Computer Society.
OpenQRM (2011). the next generation, open-
source data-center management platform.
http://www.openqrm.com/.
Pan, Q. and et al. (2008). Deep surveying of alterna-
tive splicing complexity in the human transcriptome
by high-throughput sequencing. Nature Genetics,
40(12):1413–1415.
Pratt, B. and et al. (2011). MR-Tandem: Parallel X!Tandem
using Hadoop MapReduce on Amazon Web Services.
Bioinformatics, 8:1–12.
Saaty, T. L. (1990). How to make a decision: The analytic
hierarchy process. European Journal of Operational
Research, 48(1):9 – 26. Decision making by the ana-
lytic hierarchy process: Theory and applications.
Saldanha, H. V. and et al. (2011). A cloud architecture for
bioinformatics workflows. In 1st International Con-
ference on Cloud Computing and Services Science,
CLOSER.
Schatz, M. C. (2009). CloudBurst: Highly Sensitive Read
Mapping with MapReduce. Bioinformatics, 25:1363–
1369.
Sultan, M. and et al. (2008). A Global View of Gene Ac-
tivity and Alternative Splicing by Deep Sequencing of
the Human Transcriptome. Science, 321(5891):956–
960.
Wall, D. and et al. (2010). Cloud computing for comparative
genomics. BMC Bioinformatics, 11(1):259.
Zhang, L. and et al. (2011). Gene set analysis in the cloud.
Bioinformatics, 13:1–10.
CLOSER2012-2ndInternationalConferenceonCloudComputingandServicesScience
120
