
Integrating CCD Documents
A Way towards Effective Analysis of Patients’ Health Documentation
Juha Puustjärvi
1
and Leena Puustjärvi
2
1
Department of Computer Science, University of Helsinki, P.O. Box 68, Helsinki, Finland
2
The Pharmacy of Kaivopuisto, Neitsytpolku 10, Helsinki, Finland
Keywords: HL7, Clinical Document Architecture, Electronic Health Record, Information Integration, XML-databases,
Ontologies.
Abstract: Although the original purpose of the HL7’s Continuity of Care Documents (CCD) was to deliver clinical
summaries between healthcare organizations, nowadays they are increasingly used for collecting patients’
health documentation from various healthcare providers. Usually the collected CCD documents are
organized into hierarchical structures that simplify the search of documents, e.g., grouping together the
documents by episode, clinical specialty or time period. Yet each clinical document is stored as a stand-
alone artefact, meaning that each document is complete and whole in itself. Considering each document
only as a complete and a whole in itself also has its drawback: the efficient usage of patients’ health
documentation often is data centric, meaning that data should be extracted from various documents and then
integrated according to specific criteria. Processing such queries requires the integration of the data of the
CCD documents. In this paper we present two methods for integrating CDD documents. In the first method
an XML-database is developed and the content of the documents are stored in the database. So the content
of clinical documentation can be effectively accessed by database query languages such as SQL. In the
second method an OWL ontology for CDD documents is developed and the CCD documents are
transformed in the format that is compliant with the ontology and then stored in the ontology. So the content
of clinical documentation can be easily accessed by query languages such as RQL and SPARQL. Which
integration method is appropriate depends on whether the CDD documents are based on CDA Level 2 or
CDA Level 3.
1 INTRODUCTION
An electronic medical record (EMR) is a
computerized medical record created in an
organization that delivers care, such as a hospital or
physician's office (Hartley and Jones, 2005).
Although the terms electronic medical record and
electronic health record (EHR) are commonly used
interchangeably, these terms describe completely
different concepts. EHR relate to sharing patients
health documentation (NEHTA, 2006). It relies on
functional EMRs that allow care delivery
organizations to exchange health documentation
with other care delivery organization or stakeholders
within the community, regionally, or nationally.
That is, EMRs are data sources for EHRs.
EHRs are generally assumed to be summaries
like ASTM’s Continuity of Care Record (CCR)
(CCR, 2011) or HL7’s Continuity of Care Document
(CCD) (HL7, 2007); (CCD, 2009). They may be
owned by patient or healthcare authorities. In the
former case they are usually called as personal
health records, and in the latter case they are also
called as electronic patient record archives
(Puustjärvi and Puustjärvi, 2010). Often such
archives aim to capture patients’ all health
documentation.
As defined by the ISO 13606 Reference Model
(ISO, 2012); (prEN13606, 2006), EHR systems
usually organize clinical documents into hierarchical
structures that simplify the search of documents,
e.g., grouping together the documents by episode,
clinical specialty or time period. Further, each
clinical document is stored as a stand-alone artefact,
meaning that each document is complete and whole
in itself, including context information such as who
created it, when and where and for what purposes
(Boone, 2011). Without such contextual information
in some cases it may be a risk to interpret some
values of the data included on a document.
293
Puustjärvi J. and Puustjärvi L..
Integrating CCD Documents - A Way towards Effective Analysis of Patients’ Health Documentation.
DOI: 10.5220/0004175702930300
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2013), pages 293-300
ISBN: 978-989-8565-37-2
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

Considering each document only as a complete
and a whole in itself also has its drawback. The
problem here is that the efficient usage of patients’
health documentation often is data centric, meaning
that data should be extracted from various
documents and then integrated according to specific
criteria. Especially this is a common requirement in
treating patients suffering from chronic diseases
(Michie et al., 2003); (Fiandt, 2011).
For example, a physician may be interested to
know the average blood pressure and/or cholesterol
level during the time periods the patient was using
Emconcor (a drug for blood pressure). Presenting
the context information of each document stating the
measured blood pressures is not necessary. Neither
presenting the single values of each measurement is
needed. Presenting such information may even
frustrate the physician as the physician is
overwhelmed with information.
Also the queries that access the documents of
many patients may be of prime importance. For
example, the healthcare authorities may be interested
to know the average doses of drugs that the
physicians have prescribed. Unfortunately, the
computation required by such queries is not
provided by the health information systems that are
developed for managing hierarchically organized
EHRs.
Nowadays EHR systems increasingly use HL7’s
CCD documents for collecting patients’ health
documentation although their original purpose was
to deliver clinical summaries between healthcare
organizations (Benson, 2010). Processing data
centric queries on CDD documents would be of high
importance. Yet, EHR systems do not provide this
feature as it would require the integration of the data
of the CDD documents.
In this paper we present two methods for
integrating CDD documents. In the first method, an
XML-database is developed and the content of the
documents are stored in the database. In the second
method, an ontology, called the CDD-ontology, for
the CDD documents is developed. Also the CCD
documents are transformed in the format that is
compliant with the ontology, and finally they are
stored in the ontology. Which method is appropriate
depends on whether the CDD documents are based
on CDA Level 2 or CDA Level 3.
The rest of the paper is organized as follows.
First, in Section 2, we consider the characteristics of
the clinical documents defined by the CDA standard.
In Section 2, we first consider the RIM on which the
CDA standard is based on, and then we present the
structure of CCD documents. In Section 3, we
consider the suitability of XML databases for storing
the content of CCD Level 2 documents, and in
Section 4 we consider the suitability of OWL
ontologies for storing CCD Level 3 documents.
Finally, Section 5 concludes the paper.
2 HL7 CLINICAL DOCUMENT
ARCHITECTURE
2.1 Reference Information Model
The HL7 Clinical Document Architecture (CDA) is
an XML-based markup standard intended to specify
the encoding, structure and semantics of clinical
documents for exchange (Dolin, 2001); (Boone,
2011). It is based on the HL7 Reference Information
Model (RIM) which is the UML model for
healthcare information. The idea behind the RIM is
that we can interpret the tags of the CDA documents
by the RIM.
The RIM is based on two key ideas (Benson,
2010). The first idea is based on the consideration
that most healthcare documentation is concerned
with “happenings” and things (human or other) that
participate in these happenings in various ways.
The second idea is the observation that the same
people or things can perform different roles when
participating in different types of happening, e.g., a
person may be a care provider such a physician or
the subject of care such as patient.
As a result of these ideas the RIM is based on a
simple backbone structure, involving three main
classes, Act, Role, and Entity, linked together using
three association classes Act-Relationship,
Participation, and Role-Relationship (Figure 1).
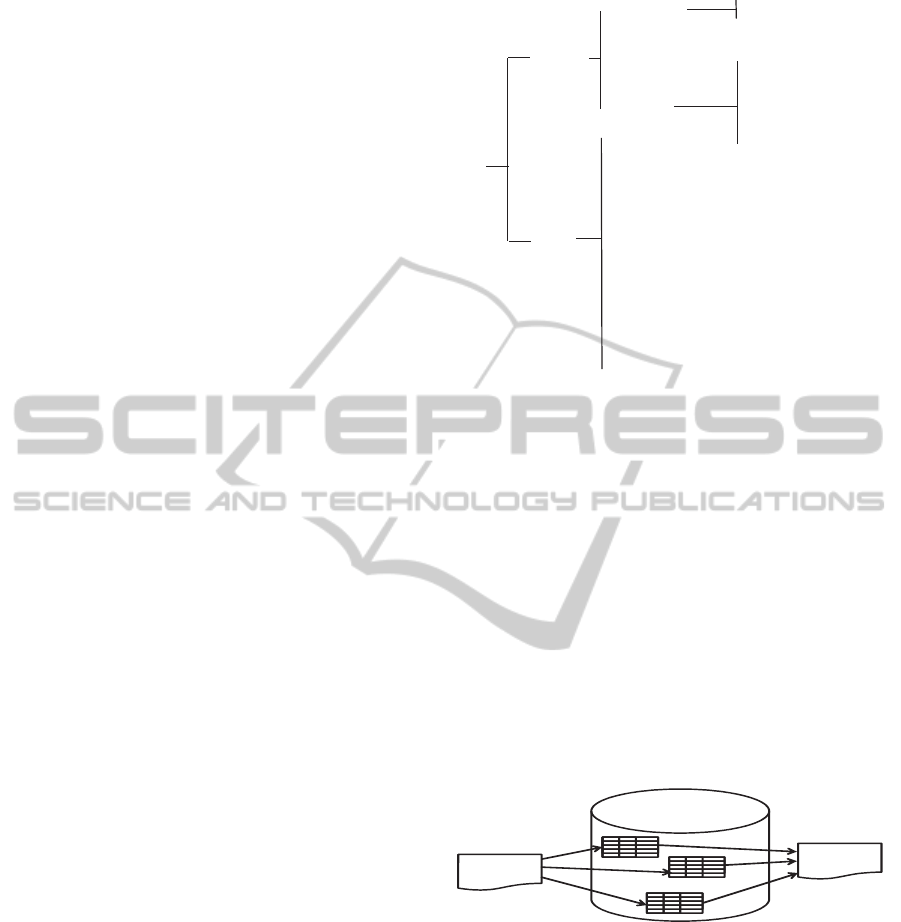
Figure 1: The RIM backbone structure.
Each happening is an Act, and it may have any
number of Participations, which are Roles, played by
Entities. An ACT may also be related to other Acts
via Act Relationships. Act, Role and Entity classes
have a number of specializations (subclasses), e.g.,
Entity has a specialization LivingSubject, which
itself has a specialization Person.
The classes in the RIM have structured attributes
which specify what each RIM class means when
HEALTHINF2013-InternationalConferenceonHealthInformatics
294
used in a message (exchanged document). The idea
behind structured attributes is to reduce the original
RIM from over 100 classes to a simple backbone of
six main classes.
Note that we cannot consider the RIM as the
ontology of the CDA documents as CDA documents
are not directly derived from the RIM but through
the constrained information models RMIMs
(Refined Message Information Model). As we will
illustrate in section 5, each RMIM diagram is
derived from the RIM by limiting its optionality
such that it specifies the semantic of a whole CDA
document or its portion.
2.2 Continuity of Care Document
The CCD (Continuity of Care Document)
specification is a constraint on the HL7 CDA
standard. The CCD standard has been endorsed by
HIMMS (Healthcare Information and Management
Systems Society Though) (HIMMS, 2013) and
HITSP (Healthcare Information Technology
Standards Panel) (HITSP, 2013) as the recommend
standard for exchange of electronic exchange of
components of health information.
Although the original purpose of the CCD
documents was to deliver clinical summaries
between healthcare organizations, nowadays it
increasingly used for other types of messages: it is
considered as set of templates because all its parts
are optional, and it is practical to mix and match the
sections that are needed (Benson, 2010). Hence,
there is a RMIM behind each CCD document.
Each CCD document has one primary purpose
(which is the reason for the generation of the
document), such as patient admission, transfer, or
inpatient discharge. A CCD document, as well as all
CDA documents, is comprised of the Header and the
Body (Benson, 2010). The sections that can appear
in the Header and in the Body in a CCD document
are presented in Figure 2.
Depending whether the header and body of the
CDA documents are based on the RIM they are
classified into three levels:
CDA Level 1: Only the header is based on the
RIM while the body is human readable text or
image.
CDA Level 2: Only the header of the document is
based on the RIM while the body is comprised of
XML coded sections.
CDA Level 3: Both the header and the body are
based on the RIM.
CCD
Header
Body
‐ DocumentID
‐ Date/timecreated
‐ Documenttype
‐ Subject(patient)
‐ Source
‐ Metadata
‐ Problems
‐ Procedures
‐ Familyhistory
‐ Socialhistory
‐ Payers
‐ Advanceddirectives
‐ Alerts
‐ Medications
‐ Immunization
‐ Medicalequipment
‐ Vitalsigns
‐ Functionalstatus
‐ Results
‐ Encounters
‐ PlanofCare
‐ UniversallyuniqueI
D
‐ OriginatorID
‐ Author
‐ Organization
‐ Language
‐ Processingstatus
‐ ConformanceID
‐ ACKrequired
Figure 2: The components of the CCD.
3 INTEGRATING CCD LEVEL 2
DOCUMENTS
3.1 XML Databases
We have exploited XML database (Obasanjo, 2001)
in integrating CCD Level 2 documents. However, in
general there are other reasons why XML documents
are stored in databases: the most common reason is
to publish data stored in a database as an XML
document. Its reverse process, i.e., extracting data
from an XML document and storing it in the
database is called shredding (Figure 3).
XMLDatabase
XML
document
XML
document
Shredding
Publishing
Figure 3: XML database.
In healthcare sector XML databases are suitable
for archiving XML-formatted EHRs in the sense that
databases are secure, and can be easily queried and
retrieved. Further, as illustrated in Figure 4, an EHR
system can be quite easily implemented by
exploiting the functionalities provided by a
document management system, which in turn
exploits the functionalities of an XML database.
Alternatively, an EHR system itself may also
provide the functionalities of the document
management system.
IntegratingCCDDocuments-AWaytowardsEffectiveAnalysisofPatients'HealthDocumentation
295

Figure 4: Archiving XML documents.
In principle, we have two choices to use XML
with databases: XML-enabled databases and native
XML databases (IBM, 2004). In the former XML is
used only as a way to exchange data. The database
schema matches the XML schema, but there is no
“XML” visible inside the database. That is, the
database includes the data items of the XML-
documents but the original XML documents cannot
be reconstructed by querying the database.
In contrast with native XML databases the
document is itself stored in the database. Thus the
structures of XML document are visible inside the
database meaning that the database contains
information such as element and attribute names. In
other words, the database schema models XML
documents, and so original XML documents can be
reconstructed by querying the database.
In our case there is no need to reconstruct
original XML-formatted CCD documents: according
to the wholeness requirement it is enough that the
content and the context of a CCD document can be
put together. Aspects like the order of the elements
in the original XML documents do not matter. This
substantially simplifies the technical requirements of
the database-based CCD archive: we just attach the
identifier of each CCD document to each tuple in the
database. Then by querying the instances by
document identifier we can return the context and
content of the CCD document. Thus the requirement
of documents human readability is also easily met.
Further the requirement of persistence can be
ensured by not allowing updates on the relations
containing the data of clinical documents. The
requirements of context and stewardship are met as
the database includes all the data included in clinical
documents. That is, if the clinical documents meet
these requirements then the documents stored in
databases also meet these requirements.
3.2 Storing CCD Documents in
XML-enabled Databases
In order to illustrate how CDD documents can be
stored in XML-enabled database consider the
simplified CCD documents presented in Figures 5
and 6.
The first document includes data gathered from
blood pressure measurements, and the second
document includes information about patient’s
medication.
Figure 5: A CCD file of blood pressures.
Figure 6: A CCD file of medication.
The representation formats of these documents in
a relational XML-enabled database are presented in
Figures 7 and 8. The basic idea behind transforming
XML documents into relation schemas is that each
complex element gives rise for a relation, and each
attribute and simple element is represented by an
attribute of the relation. Such relations are not
necessary yet in BCNF (Boyce-Codd Normal Form)
(Ullman and Widom, 1997), but their normalization
can be easily carried out by splitting the non-
normalized relations.
HEALTHINF2013-InternationalConferenceonHealthInformatics
296

Figure 7: Tuples generated from the document of Figure 5.
Figure 8: Tuples generated from the document of Figure 6.
Now, by SQL (Ullman and Widom, 1997), we
can make many useful queries on patients’ health
information. Examples of such queries include:
Patient’s average blood pressures during January
2012.
Patient’s highest blood pressure during the time
the patient was using Emconcor.
Queries that do not concern a single patient can be
also easily presented. Examples of such queries
include:
List the physicians (attribute ActorId) in the
order determined by the times they have prescribed
Emconcor.
The average blood pressures of the patients using
Emconcor (Valsartan).
Note that these queries cannot be processed by EHR
systems as they access documents by query
languages, such as XPath or XQuery (Harold and
Scott Means, 2002), which are designed for
manipulating XML documents.
4 INTEGRATING CCD LEVEL 3
DOCUMENTS
We now illustrate the development of the CDD-
ontology by integrating the RMIM ontologies, which
are derived from the RMIM diagrams of the CCD
Level 3 documents.
4.1 Developing RMIM Diagrams
The body and the header of a CDA Level 3
document are defined by RMIM diagrams. Further,
according to the HL7 Version 3 Development
Framework, the RMIM diagrams are transformer
into XML schemas, which specify the structure of
the exchanged CDA documents. In transformation
classes map to complex elements, structured
attributes map to attributes and other attributes map
to simple elements. As a result, the tags of each
CDA document represent the classes and attributes
of the RMIM. Thus, we can interpret the semantics
of the tags of the CDD documents by the RIM and
RMIM.
Each RMIM diagram is derived from the RIM by
limiting its optionality by omission and cloning
(Benson, 2010). Omission means that the RIM
classes or attributes can be left out. Note that all
classes and attributed that are not structural
attributes in the RIM are optional, and so the
designer can take only the needed classes and
attributes. Cloning means that the same RIM class
can be used many times in different ways in various
RMIMs. The classes selected for a RMIM are called
clones.
The multiplicities of associations and attributes
in a RMIM are constrained in terms of repeatability
and optionality. Further, code binding is used for
specifying the allowable values of the used
attributes.
To illustrate the relationships of the CCD, RIM
and RMIM consider the RMIM diagram of Figure 9.
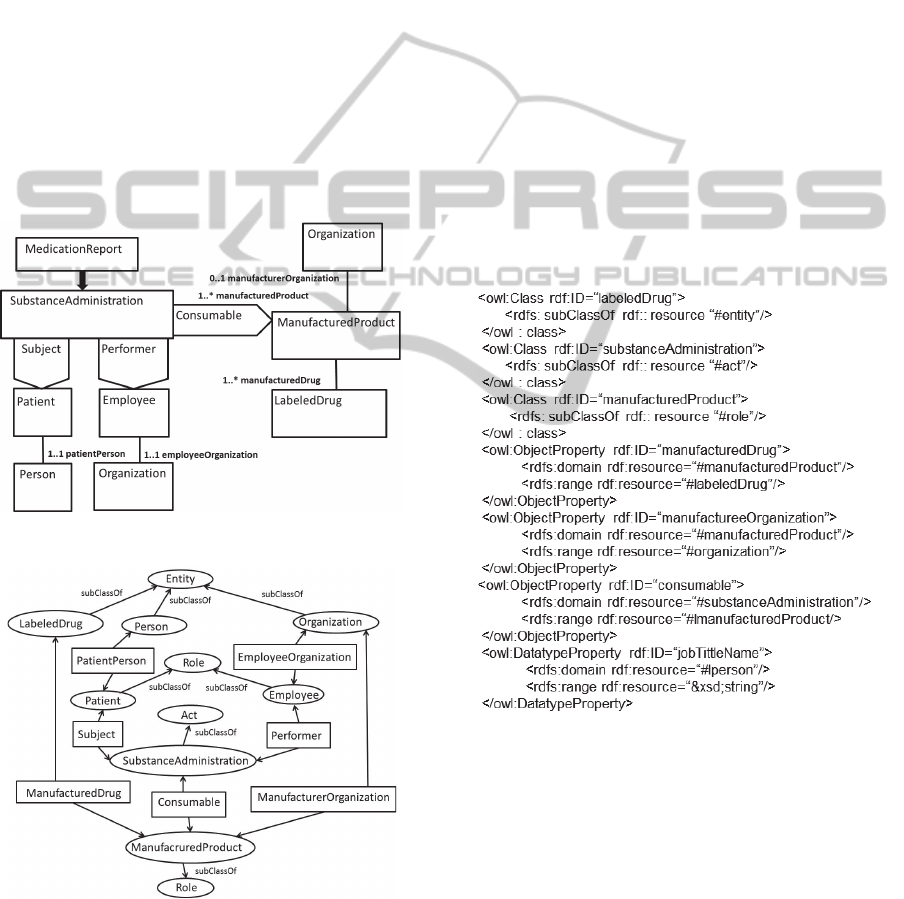
ObservationEvent
Subject Performer
Patient
Employee
Person Organization
VitalSignsEvent
ComponentOf
1..1patientPerson 1..1employeeOrganization
1..*vitalSignsEvent
BloodPressureReport
Component
BloodPressureEvent
1..*bloodpressureEvent
Figure 9: The RMIM of the blood pressure report.
Note that HL7 uses its own representation of
UML in RMIM diagrams: each class has its own
colour and shape to represent the stereotypes of
these classes, and they only connect in certain ways.
The diagram specifies a blood pressure report. Its
body includes the Vital signs section of the CCD.
The use case behind this RMIM diagram is to
IntegratingCCDDocuments-AWaytowardsEffectiveAnalysisofPatients'HealthDocumentation
297

exchange and store patient’s blood pressures
(SystolicBloodPressure and DiastolicBloodPressure)
and the time of the measurement (EffectiveTime).
These are the attributes of the clone
BloodPressureEvent but we have omitted them, as
well as other attributes, from the RMIM diagram.
The entry point of this diagram
(BloodPressureReport) is ObservationEvent, which
is specialization of the RIM class Act. Also classes
VitalSignsEvent and BloodpressureEvent are
specializations of the class Act. Classes Patient and
Employee are specializations (subclasses) of the
RIM class Role. Person and Organization are
specializations of the RIM class Entity. Subject and
Performer are specializations of the association class
Participation. Component and ComponentOf are
specializations of the association class
ActRelationship.
4.2 Transforming RMIM Diagrams
into OWL
Although the semantics of all CDA documents is
tractable through a RMIM back to the RIM, we
neither can use the RMIM nor the RIM in
formulating queries. The reason is twofold: First,
each RMIM diagram only models one type of
document. Second, there are no query languages
specified for the information model used in the
RMIM and RIM schemas.
For these reasons we first transform RMIM
diagrams into OWL (Web Ontology Language)
(OWL, 2011), and then integrate these OWL-
ontologies. The result of the integration is the CD-
ontology. As it is OWL ontology we can define data
centric queries by the query languages, such as RQL
(RQL, 2002) and SPARQL (SPARQL, 2008), which
are developed for OWL ontologies.
Transforming a RMIM diagram into OWL is
straightforward in the sense that the both models are
object-oriented although the notation used in RMIM
diagrams slightly differs from the traditional UML
notation. Yet their basic modelling primitives are the
same, namely classes, subclasses, properties and
values. The classes are also connected in a similar
way through properties.
The RMIM diagram of Figure 9 is presented in
OWL in a graphical way in Figure 10. In this
graphical representation ellipses represent classes
and subclasses while rectangles represent data type
and object properties. Classes, subclasses, data
properties and object properties are modeling
primitives in OWL (Antoniou and Harmelen, 2004).
Object properties relate objects to other objects
while datatype properties relate objects to datatype
values. Note that, in Figure 10 we have attached
datatype properties only to the class
BloodPressureEvent.
Entity
Person
Organization
Role
Patient
PatientPerson
EmployeeOrganization
Employee
Subject
Performer
ObservationEvent
ComponentOf
VitalSignsEvent
Act
subClassOf
subClassOf
subClassOf
subClassOf
subClassOf
subClassOf
Component
BloodPressureEvent
EffectiveTime
DiastolicBloodPressure
SystolicBloodPressure
Figure 10: Graphical presentation of the RMIM ontology
BloodPressureReport.
A portion of the graphical RMIM ontology of
Figure 10 is presented in OWL in Figure 11.
Figure 11: The RMIM BloodPressureReport ontology.
HEALTHINF2013-InternationalConferenceonHealthInformatics
298
4.3 Integrating RMIM Ontologies
In the development of the CDD-ontology we have
first translated a RMIM ontology into OWL. Then
this ontology (the CCD ontology) is extended step
by step by integrating other RMIM ontologies with
the ontology. Hence the CDD ontology is
incremental: when a new CDD document type
(RMIM) is introduced, the CDD-ontology is
extended accordingly.
Each integration step is comprised of two
successive phases: First, the CDD-ontology is
merged with the CDD-ontology, and then potential
conflicts are detected and resolved.
To illustrate the merging phase, consider the
CDD document (named MedicationReport), which
RMIM diagram is presented in Figure 12, and the
graphical OWL ontology derived from this RMIM
diagram is presented in Figure 13.
Figure 12: The RMIM of the medication report.
Figure 13: Graphical presentation of the RMIM ontology
MedicationReport.
In merging, we add those elements (classes,
object properties and datatype properties) from the
Medication report ontology to the Blood pressure
report ontology that do not include in both
ontologies. Such classies are
SubstanceAdministration, ManufacturedProduct,
and LabeledDrug. Correspondingly such object
properties are Consumable, ManufacturedProduct
and ManufacturedOrganization.
Note that in graphical OWL representations (for
simplicity) we have specified only a few datatype
properties (only the class BloodPressureEvent has
attached datatype properties in Figure 10), and so
our used examples do not reveal the datatype
properties that we should insert in the integrated
ontology (CDD ontology).
However, assuming that clone (class) Person has
the datatype property JobTitleName in the
Medication report ontology but not in the Blood
Pressure report ontology, then the datatype property
JobTitleName should be inserted into the integrated
ontology. So, in the merging phase we have to insert
the OWL code of Figure 14 to the ontology
presented in Figure 11.
Figure 14: The OWL code to be inserted in merging the
Blood pressure report and Medication report ontologies.
After this we have to detect and resolve conflicts.
However, in the context of RMIM ontologies
detecting and resolving conflicts is not as complex
as in general: the “backbone structure” of the RIM
ensures that the same concept has the same
semantics in all RMIM ontologies. The only sources
of heterogeneity arise from constraining the classes
(clones) in different ways.
IntegratingCCDDocuments-AWaytowardsEffectiveAnalysisofPatients'HealthDocumentation
299

4.4 CCD Level 3 Documents
Storing CCD Level 3 documents into the CDD-
ontology requires that they are first transformed (by
an XSLT-based style sheet engine (Harold and
Scott, 2002) into RDF/XML format that is compliant
with the CDD-ontology. The transformation requires
that we have to define a stylesheet for each type of
CDD document (Figure 15).
Reconstructing the content of original documents
(or representing queries) by RQL and SPARQL on
the CDD-ontology is rather easy. For example, in
RQL to retrieve all instances of the class
BloodPressureEvent (i.e., all measured blood
pressures) we only have to write
“BloodPressureEvent”. However, the physicians do
not have to be familiarized with query languages in
order to retrieve data from the CDD-ontology as
user-friendly interfaces can be easily developed.
5 CONCLUSIONS
The Clinical Document Architecture (CDA) is
proven to be a valuable and powerful standard for a
structured exchange of clinical documents between
healthcare information systems.
What is still missing is the conceptual model of
patient’s health data. Without a conceptual schema
we can neither query health data nor can we store
health data in a way which allows data centric
queries, and therefore we have focused on this
problem. We have shown how the integration can be
carried out by exploiting XML-enabled relational
databases if the CCD documents are based on the
CDA Level 2. Further, we have shown how
ontology-based integration can be carried out if the
CCD documents are based on the CDA Level 3. Still
an open problem is how to integrate CCD Level 2
and CCD Level 3 documents among themselves.
This is an issue of our future work.
REFERENCES
Antoniou, G., Harmelen, F., 2004. A Semantic Web
Primer. The Mitt Press.
Benson, T., 2010. Principles of Health Interoperability
HL7 and SNOMED. Springer.
Boone, K., 2011. The CDA Book. Springer.
CCD, 2009. What Is the HL7 Continuity of Care
Document? Available at: http://www.neotool.com/
blog/2007/02/15/what-is-hl7-continuity-of-care-docu
ment/
CCR, 2011. Continuity of Care Record (CCR) Standard.
Available at: http://www.ccrstandard.com/
Dolin, R., Alschuler, L., Beerb, C., Biron, P., Boyer, S.,
Essin, E., Kimber, T. 2001. The HL7 Clinical
Document Architecture. J. Am Med Inform Assoc,
2001:8(6), pp. 552-569.
Fiandt, K., 2011. The Chronic Care Model: Description
and Application for Practice. Available at:
http://www.medscape.com/viewarticle/549040
Harold, E., Scott Means, W., 2002. XML in a Nutshell.
O’Reilly & Associates.
Hartley, C., Jones, E., 2005. EHR Implementation. AMA
Press.
HIMMS, 2013. Healthcare Information and Management
Systems Society, Available at: http://himms.org/
HITSP, 2013. Healthcare Information Technology
Standards Panel, Available at: http://hitsp.org/
HL7, 2007. What is the HL7 Continuity of Care
Document? Available at:
http://www.neotool.com/blog/2007/02/15/what-is-hl7-
continuity-of-care-document/
IBM, 2004. XML for DB2 Information Integration.
Available at: http://www.redbooks.ibm.com/
redbooks/pdfs/sg246994.pdf.
ISO, 2012. 13606 Electronic Health Record
Communication. Available at:
http://discovery.ucl.ac.uk/66213/
Michie, S., Miles, J., Weinman, J., 2003. Patient-
centredness in chronic illness: what is it and does it
matter?. Patient Education and Counselling, pp. 197-
206.
NEHTA. 2006. Review of shared electronic health record
standards. Version 1.0. National e- Health Transition
Authority, Available at: http://www.nehta.gov.au/
component/option,com_docman/task,cat_view/gid,130
/Itemid,139/
Obasanjo, D., 2001. An Exploration of XML in Database
Management Systems. Available at: http://
www.25hoursaday.com/StoringAndQueryingXML.ht
ml
OWL, 2011. WEB OntologyLanguage. Available at:
http://www.w3.org/TR/owl-features/
prEN13606, 2006.Health informatics – Electronic
healthcare record communication – Parts 1-5.
Committee European Normalisation, CEN/TC 251
Health Informatics Technical Committee. Available at:
http://www.centc251.org/
Puustjärvi, J., Puustjärvi, L., 2010. Automating the
Importation of Medication Data into Personal Health
Records. In the proc. of the International Conference
on Health Informatics. Pages 135-141.
RQL, 2002. RQL: A Declarative Query Language for
RDF, Available at: http://www2002.org/
CDROM/refereed/329/
SPARQL, 2008. SPARQL Query Language for RDF.
Available at: http://www.w3.org/TR/rdf-sparql-query/
Ullman, D., Widom, J., 1997. A First Course in Database
Systems. Prentice Hall.
HEALTHINF2013-InternationalConferenceonHealthInformatics
300
