
Performance of Beta-Binomial SGoF Multitesting Method for
Dependent Gene Expression Levels
A Simulation Study
Irene Castro-Conde
1
and Jacobo de Uña-Álvarez
1,2
1
SiDOR Research Group, University of Vigo, Facultad de Económicas, 36310 Vigo, Spain
2
Department of Statistics and OR, University of Vigo, Facultad de Económicas, 36310 Vigo, Spain
Keywords: Correlated Tests, False Discovery Rate, Gene Expression Levels, Monte Carlo Simulations, Multitesting
Procedures.
Abstract: In a recent paper (de Uña-Álvarez, 2012, Statistical Applications in Genetics and Molecular Biology Vol.
11, Iss. 3, Article 14) a correction of SGoF multitesting method for possibly dependent tests was introduced.
This correction enhanced the field of applications of SGoF methodology, initially restricted to the
independent setting, to make decisions on which genes are differently expressed in group comparison when
the gene expression levels are correlated. In this work we investigate through an intensive Monte Carlo
simulation study the performance of that correction, called BB-SGoF (from Beta-Binomial), in practical
settings. In the simulations, gene expression levels are correlated inside a number of blocks, while the
blocks are independent. Different number of blocks, within-block correlation values, proportion of true
effects, and effect levels are considered. The allocation of the true effects is taken to be random. False
discovery rate, power, and conservativeness of the method with respect to the number of existing effects
with p-values below the given significance threshold are computed along the Monte Carlo trials.
Comparison to the classical Benjamini-Hochberg adjustment is provided. Conclusions from the simulation
study and practical recommendations are reported.
1 INTRODUCTION
Multiple-testing problems have received much
attention since the advent of the –omic technologies:
genomics, transcriptomics, proteomics, etc. They
often involve the simultaneous testing of hundreds
or thousands of hypotheses, or nulls, producing as a
result a number of significant p-values or effects
(that is, an increase in gene expression, or
RNA/protein levels). In this setup, the family-wise
error rate (FWER) and the false discovery rate
(FDR) have been proposed as suitable significance
criteria to perform the multiple testing. See
Benjamini and Hochberg (1995), Nichols and
Hayasaka (2003) or Dudoit and Laan (2008) for
basic definitions and reviews of existing literature.
As a drawback of FWER-based and FDR-based
methods, their power may be small when the number
of tests is large, or when the proportion of true nulls
is large and the effect in the non-true nulls is weak
relative to the sample size (Carvajal-Rodríguez et
al., 2009); (de Uña-Álvarez, 2011). Carvajal-
Rodríguez et al., (2009) introduced a new
multitesting strategy, SGoF (from Sequential
Goodness-of-Fit), which focuses on the difference
between the observed proportion of p-values below a
given significance threshold (the γ parameter) and
the expected one under the complete null of no
effects (γ); therefore, a binomial test for a proportion
is performed. SGoF approach provides a reasonable
compromise between false discoveries and power
(Carvajal-Rodríguez et al., 2009); the theoretical
statistical properties of SGoF were investigated in
detail in de Uña-Álvarez (2011). It was illustrated
that, with a large number of tests, the critical region
provided by SGoF is wider than that of FDR-based
methods in most practical scenarios. SGoF original
method provides reliable inference when the
multiple tests are independent.
However, in real world applications,
dependences among the tests will appear. This
dependence may be provoked by the existence of
several blocks of tests which share the same within-
93
Castro-Conde I. and de Uña-Álvarez J..
Performance of Beta-Binomial SGoF Multitesting Method for Dependent Gene Expression Levels - A Simulation Study.
DOI: 10.5220/0004191100930097
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2013), pages 93-97
ISBN: 978-989-8565-35-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

block probability of reporting a significant p-value.
While FDR-based strategies are robust in
dependence scenarios, the same is not true for SGoF,
which crucially depends on the correct estimation of
the variance associated to the number of discoveries.
In most practical situations with dependent tests, the
final number of discoveries reported by SGoF will
be too liberal, because it will be based on an
underestimated variance (Owen, 2005). To solve this
issue, de Uña-Álvarez (2012) introduced a
correction of SGoF method to deal with dependent
tests. This correction is based on the beta-binomial
extension of the binomial model, which arises when
the number of successes S among the n trials is
conditionally distributed as a binomial given the
probability of success π, which is a random variate
following a beta distribution. The beta-binomial
model has three parameters: the number of trials n,
the mean probability of success p=E(π), and the
pairwise correlation between the outcomes
τ=Var(π)/p(1-p). The mean and the variance of the
beta-binomial model are given respectively by
E(S)=np and Var(S)=np(1-p)(1+(n-1)τ); this shows
that, by putting τ>0, the beta-binomial model allows
for a variance larger than binomial. See Johnson and
Kotz (1970) for further details and illustrations of
the model.
More specifically, given the set of n p-values
u
1
,…,u
n
coming from the n nulls being tested, BB-
SGoF (from beta-binomial SGoF) correction starts
by computing the binary sequence X
i
=I(u
i
≤γ),
i=1,…,n. Then, by assuming that there are k
independent blocks of p-values of sizes n
1
,…,n
k
(with n
1
+…+n
k
=n), the number of successes sj
within each block j is computed. Here, X
i
=1 is called
‘success’. Each s
j
is assumed to be a realization of a
beta-binomial variable with parameters (n
j
,p,τ). In
this setting, p represents the average proportion of p-
values falling below γ, which under the complete
null is just γ, and τ is the within-block correlation
between two outcomes X
i
and X
j
. Estimation of p
and τ is performed by maximum-likelihood, and the
lower bound of a 100(1-α)% interval for the excess
of significant cases n(p-γ) is reported; this bound is
the number of effects declared by BB-SGoF (which
weakly controls FWER at level α in this manner).
Therefore, BB-SGoF follows the same spirit of
SGoF method, but some preliminary estimation of
the p-values’ dependence structure is performed to
correct for it. This correction may have a big impact
in the researcher’s decision; for example, de Uña-
Álvarez (2012) illustrates for two real datasets that
ordinary SGoF rejects 10% (Hedenfalk data) or
about 4% (Diz data) nulls more than BB-SGoF, and
that BB-SGoF rather than SGoF should be applied
due to significant correlation.
Simulations in de Uña-Álvarez (2012) for n=500
and n=1000 tests reported the mean and standard
deviation of the number of rejections for SGoF-type
methods, as well as the family-wise rejection rate
(which reduces to the FWER and FDR under the
complete null of no effects); these simulation results
showed that BB-SGoF is able to control FWER at
level α even when the number of blocks k is
unknown (which is the usual situation in practice),
provided that some conservative criterion in the
estimation of k is used. Besides, this conservative
criterion did not result in a great loss of power
(compared to the ‘benchmark’ method based on the
true value of k). Ordinary SGoF performed badly
otherwise, being unable to control for dependences
(as expected). However, the simulation study
reported in the referred paper has some limitations.
First, no computation of the FDR in the presence of
effects was made, neither results on the methods’
power were reported. This was because of the
employed simulation procedure, which does not
allow for distinguishing between true and non-true
nulls. Also because of this, comparison to the
Benjamini-Hochberg (BH) FDR-controlling
procedure was not possible. Second, the simulated
model was a beta-binomial, and therefore the
simulations are useless to know how the beta-
binomial approach will work in other scenarios with
blocks of dependent tests. The simulation study in
the present work aims to overcome these limitations.
The rest of the paper is organized as follows. In
Section 2 we describe the simulated setting. In
Section 3 we report the results of our simulation
study, and we summarized them in a number of
relevant findings. A final discussion is reported in
Section 4.
2 SIMULATED SETTING
Having in mind the study of Hedenfalk data (see e.g.
de Uña-Álvarez, 2012), we simulated n=500 or
n=1000 2-sample t-tests for comparison of normally
distributed ‘gene expression levels’ in two groups A
and B with sizes 7 and 8 respectively. The
proportion of true nulls (i.e. genes equally
expressed) Π
0
was 1 (complete null), 0.9 (10% of
effects), or 0.67 (33% of effects). Mean was always
taken as zero in group A, while in group B it was μ
for 1/3 of the effects and –μ for the other 2/3 of
effects, with μ=1 (weak effects), μ=2 (intermediate
effects), or μ=4 (strong effects). Random allocation
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
94
of the effects among the n tests (genes) was
considered. Within-block correlation levels of ρ=0,
0.1, 0.2 and 0.8 were taken. With regard to the
number of blocks, we considered k=10 or k=20, so
we had 50 or 25 tests per block when n=500, and
100 or 50 tests per block when n=1000. For random
generation, the function rmvnorm of the R software
was used.
BB-SGoF method with γ=α=0.05 was applied
under perfect knowledge on the true value of k but
also when underestimating (k/2) or overestimating
(2k) the true number of blocks. The blocks were
always constructed homogeneous in size. We also
applied an automatic (data-driven) choice of k by
minimizing the number of effects declared by BB-
SGoF along the grid k=2,…,61. For γ=0.05, 5% of
the p-values are expected to fall below γ under the
complete null, while this amount rises to about
17.8%, 34.9% and 36.7% when e.g. there is a 33%
of weak, intermediate, or strong effects respectively.
The estimated values of the within-block correlation
τ between indicators X
i
=I(u
i
≤γ) and X
j
=I(u
j
≤γ) in the
simulations were much smaller than the within-
block correlation ρ between the ‘gene expression
levels’, ranging from about 0.002 to 0.232
depending on ρ.
1000 Monte Carlo simulations were performed.
For each situation, we computed the FDR, the power
(both averaged along the 1000 trials), and the
proportion of trials for which the number of declared
effects was not larger than the number of effects
with p-value below γ (this is just 1-FDR under the
complete null); as indicated in de Uña-Álvarez
(2012), BB-SGoF guarantees that this proportion
(labeled as Cov from ‘coverage’ in Tables below) is
asymptotically (i.e. n→∞) larger than or equal to 1-
α, a property which is not shared by other
multitesting methods. Computation of these
quantities for the original SGoF method for
independent tests and for the BH method (with a
nominal FDR of 5%) was also included to compare.
3 SIMULATION RESULTS
Tables 1 to 6 reported in this section are a sample of
the full set of results of our simulations. They are
restricted to case k=10, n=1000, no effects or 33%
of effects, weak or strong effects (μ=1 or μ=4), and
within-block correlation ρ=0 (independent setting),
ρ=0.2 (moderate correlation), and ρ=0.8 (strong
correlation). Other cases reported similar results (not
shown).
Starting with the case of no effects, we see from
Tables 1, 3 and 5 that all the methods respect the
nominal FWER (FDR) of 5% fairly well in the
independent setting. The automatic BB-SGoF
reports an FDR below nominal, something expected
due to its conservativeness. As correlation grows,
original SGoF for independent tests loses control of
FWER; when ρ=0.8, it is almost 7 times the
nominal. On the other hand, BB-SGoF methods
adapt well to the correlated settings (particularly true
for the benchmark method which uses the true k, and
for the automatic method), while BH method
respects the nominal FDR (expected, due to its
robustness for dependences) but it is very
conservative in the case ρ=0.8. The FWER of BB-
SGoF is above the nominal when the researcher
overestimates the number of blocks; this is because
BB-SGoF decision becomes more liberal as the
assumed dependence structure gets weaker.
Summarizing, the results for BB-SGoF are relevant
since they suggest FWER control even when the
simulated model is not beta-binomial.
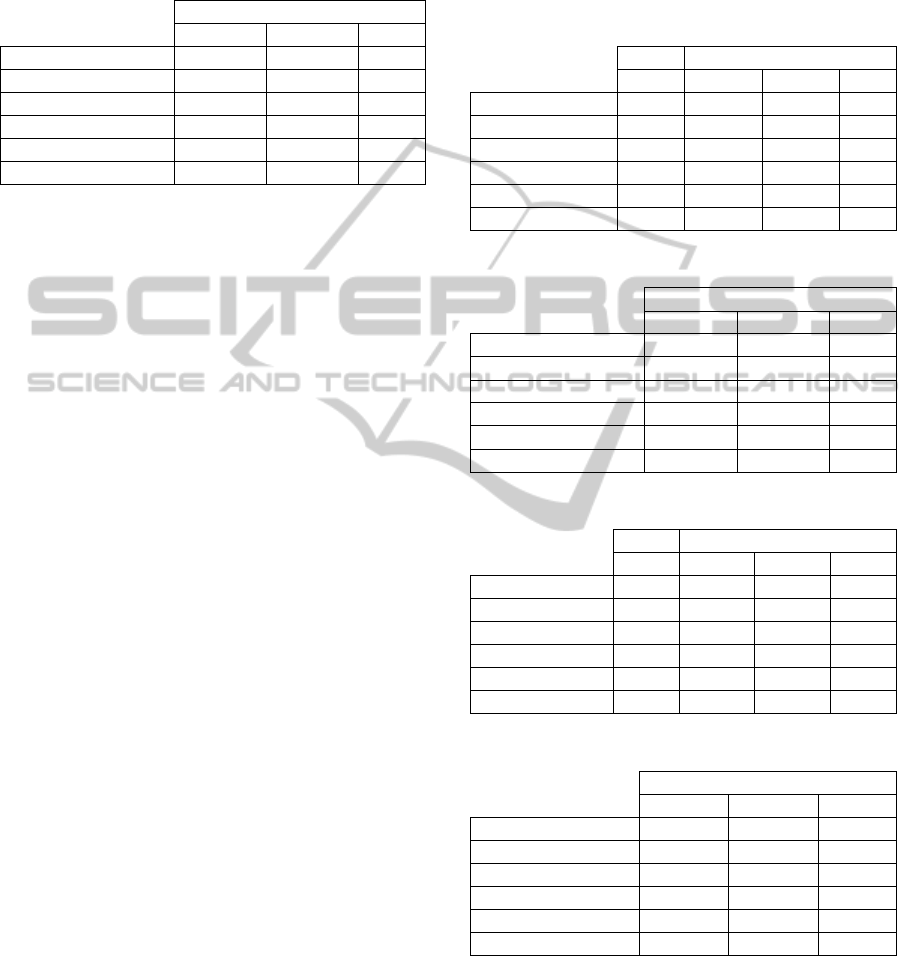
Table 1: n=1000, ρ=0, k=10, μ =1 (see text).
Π
0
=1 Π
0
=0.67
FDR FDR Power Cov
SGoF
0.048 0.1260 0.2852 1
BH
0.057 0.0353 0.0301 1
BB-SGoF (k)
0.047 0.1246 0.2808 1
BB-SGoF (k/2)
0.044 0.1239 0.2790 1
BB-SGoF (2k)
0.044 0.1250 0.2821 1
Auto BB-SGoF
0.019 0.1193 0.2679 1
The situation with 33% of weak effects (μ=1,
Tables 1, 3 and 5) reveals that SGoF-type strategies
are not controlling FDR at any given level. For
example, in the independent setting, original SGoF
and benchmark BB-SGoF report a FDR of 12.6%
and 12.5% respectively, two times and a half the
nominal FDR for BH procedure. Results for the
dependent setting are of the same order, although for
strong correlation (ρ=0.8) these FDRs go down to
10.9% and 8.5% respectively. However, the
proportion of true effects detected by SGoF-type
methods is between 5 and 9 times that of BH, the
relative performance of SGoF getting better as
correlation decreases. At the same time, one may say
that BB-SGoF is not detecting ‘too many effects’ in
the sense that, in at least 98.1% of the trials (worst
situation), the number of declared effects is below
the number of true effects with p-value below γ. It is
not strange that this proportion is just 100% for BH
since this method is rejecting only between 3% and
4% of the existing effects. Interestingly, automatic
BB-SGoF does not lose much power to respect to its
PerformanceofBeta-BinomialSGoFMultitestingMethodforDependentGeneExpressionLevels-ASimulationStudy
95
optimal version based on the true number of blocks:
its power is 6.4% smaller in the worse situation
(ρ=0.8).
Table 2: n=1000, ρ=0, k=10, μ =4 (see text).
Π
0
=0.67
FDR Power Cov
SGoF
0.0001 0.8779 1
BH
0.0333 1.0000 0
BB-SGoF (k)
0.0001 0.8693 1
BB-SGoF (k/2)
0.0001 0.8662 1
BB-SGoF (2k)
0.0001 0.8717 1
Auto BB-SGoF
0.0001 0.8533 1
The case with 33% of strong effects (μ=4, Tables
2, 4 and 6) allows to see that, in some instances, the
FDR of SGoF-type methods may be very small
compared to γ (the p-value threshold) or α (the
FWER-controlling parameter under the complete
null). Tables indicate that, for the simulated settings,
the average proportion of false discoveries of
benchmark BB-SGoF lies between 0.07/1000 and
0.4/1000, being even smaller for its automatic
version. The reason for this is that, with such strong
effects, the non-true nulls report very small p-values,
which are clearly separated from those of the true
nulls. Still, automatic BB-SGoF is able to detect
more than 80% of the existing effects. The power of
BH procedure is larger than that, according to its
higher FDR; indeed, this power is almost 100%.
This situation may be regarded as non-optimal
however, in the sense of the coverage; for example,
in the case ρ=0.8 (Table 6), only for 17% of the
1000 Monte Carlo trials the number of effects
declared by BH was below the true number of
effects with p-value smaller than 0.05, showing an
anticonservative performance in this sense (this
percentage was even smaller for the other correlation
levels). Also importantly, as for the case with weak
effects, the automatic choice of the number of blocks
results in a small loss of power (smaller than 2.5% in
this case).
The number of blocks of dependent tests
detected by automatic BB-SGoF was not always
close to the true k (k=10). For example, under the
complete null it was 18.1 (independent setting), 10.4
(ρ=0.2), or 6.9 (ρ=0.8) on average, therefore being
decreasing with an increasing correlation.
Corresponding standard deviations were 16.7, 12.4,
and 10.5, showing a large variability of the selected
number of blocks along replicates. The average
number of blocks detected was decreasing for an
increasing proportion of effects and also for more
distant alternatives (stronger effects). Whatever the
case, one should keep in mind that the role of
automatic BB-SGoF is not to perfectly estimate the
number of existing blocks but rather to allow for
error control in the multitesting procedure when the
value of k is unknown.
Table 3: n=1000, ρ=0.2, k=10, μ =1 (see text).
Π
0
=1 Π
0
=0.67
FDR FDR Power Cov
SGoF
0.145 0.1277 0.2849 1
BH
0.05 0.0302 0.0305 1
BB-SGoF (k)
0.064 0.1238 0.2745 1
BB-SGoF (k/2)
0.077 0.1238 0.2745 1
BB-SGoF (2k)
0.092 0.1251 0.2785 1
Auto BB-SGoF
0.042 0.1202 0.2634 1
Table 4: n=1000, ρ=0.2, k=10, μ =4 (see text).
Π
0
=0.67
FDR Power Cov
SGoF
0.0001 0.8783 1
BH
0.0334 1 0
BB-SGoF (k)
0.0001 0.8674 1
BB-SGoF (k/2)
0.0001 0.8650 1
BB-SGoF (2k)
0.0001 0.8707 1
Auto BB-SGoF
0.0001 0.8503 1
Table 5: n=1000, ρ=0.8, k=10, μ =1 (see text).
Π
0
=1 Π
0
=0.67
FDR FDR Power Cov
SGoF
0.341 0.1088 0.2813 0.886
BH
0.028 0.0263 0.0409 1
BB-SGoF (k)
0.059 0.0847 0.2195 0.992
BB-SGoF (k/2)
0.049 0.0857 0.2257 0.995
BB-SGoF (2k)
0.118 0.0936 0.2434 0.981
Auto BB-SGoF
0.024 0.0778 0.2054 0.999
Table 6: n=1000, ρ=0.8, k=10, μ =4 (see text).
Π
0
=0.67
FDR Power Cov
SGoF
0.0050 0.8708 0.91
BH
0.0320 0.9999 0.17
BB-SGoF (k)
0.0004 0.8258 0.995
BB-SGoF (k/2)
0.0003 0.824 0.996
BB-SGoF (2k)
0.0012 0.8458 0.982
Auto BB-SGoF
0.0001 0.8053 1
We end this section by mentioning that the
simulations with n=500 tests or with k=20 blocks
reported basically the same results as those provided
in Tables 1 to 6. However, important differences
were seen when considering a smaller number of
effects (10% instead of 33%) or intermediate effects
(μ=2) rather than weak (μ=1) or strong (μ=4) effects.
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
96

For example, with 10% of weak effects, the power
of automatic BB-SGoF relative to BH was above 17
under independence and above 15 with ρ=0.2, being
also true that BB-SGoF showed a FDR more than
twice its value with 33% of weak effects. On the
other hand, with 33% of intermediate effects, BH
and BB-SGoF procedures performed similarly in
FDR and power.
4 DISCUSSION
In this work we have investigated through
simulations the performance of BB-SGoF method.
Rate of false discoveries (FDR), proportion of
detected effects (power), and conservativeness with
respect to the true number of effects with p-value
smaller than the given threshold have been
computed. One conclusion of our research is that
BB-SGoF method may control for FWER in the
weak sense even when the underlying model is not
beta-binomial. BB-SGoF method is also robust with
respect to miss-specification of the number of
existing blocks, although it becomes too liberal
when this parameter is overestimated. As a
compromise, the automatic BB-SGoF procedure
introduced in de Uña-Álvarez (2012) performs well,
with only a small loss of power with respect to the
benchmark version. Summaryzing, BB-SGoF is a
correction of SGoF method with a suitable error
control in the presence of dependent tests; its
advantages over classical FDR-controlling strategies
(e.g. the BH method) remain the same in the
dependence scenario as for SGoF in the independent
setting, these are: greater power in the case of large
number of tests and small to moderate number of
weak effects. In such cases application of BB-SGoF
is recommended due to its compromise between
FDR and power.
ACKNOWLEDGEMENTS
Work supported by the Grants MTM2008-03129
and MTM2011-23204 (FEDER support included) of
the Spanish Ministry of Science and Innovation.
Support from the Xunta de Galicia Grant
10PXIB300068PR is also acknowledged. This work
was also supported by Agrupamento INBIOMED,
2012/273, from DXPCTSUG-FEDER ‘Unha
maneira de facer Europa’.
REFERENCES
Benjamini Y., Hochberg Y., 1995. Controlling the False
Discovery Rate: A Practical and Powerful Approach to
Multiple Testing. Journal of the Royal Statistical
Society Series B (Methodological) 57, 289-300.
Carvajal-Rodríguez A., de Una-Álvarez J., Rolan-Alvarez
E., 2009. A new multitest correction (SGoF) that
increases its statistical power when increasing the
number of tests. BMC Bioinformatics 10:209.
de Uña-Álvarez J., 2011. On the statistical properties of
SGoF multitesting method. Statistical Applications in
Genetics and Molecular Biology Vol. 10, Issue 1,
Article 18.
de Uña-Álvarez J., 2012. The beta-binomial SGoF method
for multiple dependent tests. Statistical Applications in
Genetics and Molecular Biology Vol. 11, Issue 3,
Article 14.
Dudoit S., van der Laan M., 2008. Multiple Testing
Procedures with Applications to Genomics. New
York: Springer.
Johnson N. L., Kotz, 1970. Distributions in statistics,
continuous univariate distributions-2. Houghton
Mifflin, Boston.
Nichols T., Hayasaka S., 2003. Controlling the familywise
error rate in functional neuroimaging: a comparative
review. Statistical Methods in Medical Research 12,
419-446.
Owen A., 2005. Variance of the number of false
discoveries. Journal of the Royal Statistical Society
Series B-Statistical Methodology 67, 411-426.
PerformanceofBeta-BinomialSGoFMultitestingMethodforDependentGeneExpressionLevels-ASimulationStudy
97
