
Identifying Most Relevant Concepts to Describe
Clinical Trial Eligibility Criteria
Krystyna Milian
1,2
, Anca Bucur
2
, Frank van Harmelen
1
and Annette ten Teije
1
1
VU University Amsterdam, Amsterdam, The Netherlands
2
Philips Research Eindhoven, Eindhoven, The Netherlands
Keywords:
Semantic Analysis, Selecting Ontology Subsets, Concepts Relevance, Ontology Annotators, Medical Ontolo-
gies.
Abstract:
Since eligibility criteria of clinical trials are represented as free text, their automatic interpretation and the eval-
uation of patient eligibility is challenging. Our approach to the criteria processing is based on the identification
of contextual patterns and semantic concepts that together define the machine-interpretable meaning. The goal
of this research is to find the most relevant concepts occurring in eligibility criteria that need to be mapped
to patient record to enable automatic evaluation of patient eligibility. Based on the analysis of annotation of
breast cancer trials obtained using different concept recognizers and ontologies from UMLS Thesaurus, we
chose to use MetaMap and SNOMED CT to create the mapping set. To prioritize the identified concepts, we
used the tf-idf measure and the corpus of over 38, 000 various clinical trials, to detect concepts specific for
breast cancer, and cancer in general. The obtained results can guide the mapping order of criteria concepts to
patient data. The observed substantial overlap between the terms occurring in criteria from the trials related to
breast cancer and other diseases will reduce the cost of extending the trial matching system to other diseases.
1 INTRODUCTION
Clinical trials examine the efficacy of diagnosis and
treatment methods through case-control studies, but
finding eligible patients is expensive and difficult. A
patient is enrolled in a clinical trial only when all
the eligibility criteria are fulfilled. They regard i.a.
age, gender, the current and prior diagnoses and treat-
ments. The problem is that they are defined in free
text e.g. ’No prior cancer except for skin cancer’. In
our previous work we built the patterns that capture
general meaning of criteria (e.g. ’No prior [] except
[]’) which, when detected, provide crucial context in-
formation (Milian et al., 2012). Here, we explore the
concepts that occur in eligibility criteria related to a
particular disease. Identified concepts will be used to
link to corresponding data items in patient record, to
enable evaluation of patient eligibility. The links can
be defined via the pointers to the type of a source doc-
ument (e.g. pathology report, discharge summary),
and/ or by defining semantic relations (isA, sameAs)
to the terminology locally used in a hospital. Such
process will require significant manual effort, during
the design or evaluation and involvement of medical
experts. Since medical ontologies contain hundreds
of thousands of concepts, there is a need to extract
subsets which are relevant for a particular purpose.
This study presents the experiment conducted to
compare 2 major ontology annotators: Bioportal and
MetaMap, and coverage of criteria from ClinicalTri-
als.gov
1
by the various medical ontologies (section
2). Further, section 3 describes in detail the MetaMap
annotation results of eligibility criteria of breast can-
cer trials, the quantitative characteristics of identified
concepts, their distribution over semantic types and
analysis of stability of obtained set. Section 4 demon-
strates the strategy used to prioritize the detected con-
cepts for creating mappings to patient record, and
presents findings about overlap of concepts occurring
in various types of trials. Final sections presents re-
lated work and conclusions.
2 DEFINING A STRATEGY
2.1 Selecting an Ontology Annotator
There are two major concepts recognizers available
for biomedical text mining: MetaMap (Aronson and
Lang, 2010) and NCBO annotator (Musen et al.,
2008). This section presents the experiment con-
ducted to compare the results of annotation of both
1
http://clinicaltrials.gov/
161
Milian K., Bucur A., van Harmelen F. and ten Teije A..
Identifying Most Relevant Concepts to Describe Clinical Trial Eligibility Criteria.
DOI: 10.5220/0004192501610166
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2013), pages 161-166
ISBN: 978-989-8565-37-2
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

tools on the trials corpus. We used 2135 trials from
ClinicalTrials.gov, related only to breast cancer, as it
is our main domain of interest. Both tools are highly
configurable, allow i.a. to select ontologies used for
annotations, MetaMap - any from UMLS (which inte-
grates more than 100 vocabularies), Bioportal - 16 out
of them. Because SNOMED CT is the largest relevant
ontology covered by both tools, we performed the ex-
periment restricting the vocabulary source to this one.
MetaMap returns the UMLS identifiers (CUI) of de-
tected concepts, Bioportal - the codes from a local on-
tology. To compare the results, we used the UMLS
API to retrieve the corresponding CUIs of SNOMED
ConceptIds, returned by NCBO annotator.
Figure 1 presents the annotation results, the num-
ber of concepts recognized by both tools, the over-
lap between them and the number of concepts found
only by one of them. Initially NCBO returned 7081
Figure 1: The number of SNOMED CT concepts detected
by Bioportal and MetaMap in the corpus of eligibility crite-
ria from 2135 breast cancer trials.
distinct concepts, which were mapped to 5005 CUIs.
The inspection of some of the remaining ones showed
that they were flagged in UMLS as duplicate or am-
biguous. As can be seen in the figure, MetaMap re-
turned a larger number of concepts (5994 vs 5005).
The overlap between both results is 4075, meaning
only 59% of entire set (6924), which is rather worri-
some. Additionally, both tools detected concepts not
found by the other (1919 - MetaMap, 930 - Bioportal).
Table 1 provides the details about the top 3 semantic
types present in the set of concepts detected exclu-
sively by one of the tool. In the set detected only by
MetaMap the majority of concepts have types: Find-
ings, Disease or Syndrome, Laboratory procedure.
Third on the list of Bioportal is Therapeutic or Pre-
ventive procedure. In most cases, except for Diseases,
MetaMap returned more concepts.
The proper evaluation should also consider preci-
sion and recall of both tools. However, it would re-
quire the involvement of domain experts, which ex-
ceeds the scope of this work. In paper (Shah et al.,
2009) the authors of Bioportal report that Mgrep
(algorithm underlying the service) has higher preci-
sion than MetaMap when detecting UMLS concepts
having ’Disease or syndrom’ semantic type (0.87 vs
Table 1: Number of concepts belonging to particular seman-
tic types, detected only by MetaMap and only by NCBO,
percentage of all of a type, detected by corresponding tool.
Sementic Type Only by
MetaMap
Only by
NCBO
Finding 185 (43%) 139 (22%)
Disease or Syndrome 151 (26%) 189 (49%)
Laboratory Procedure 100 (59%) 31 (11%)
Therapeutic or Preven-
tive Procedure
95 (31%) 55 (21%)
0.71), but lower in case of ’Biological processes’ (0.6
vs 0.63). Clearly, the performance is dictionary de-
pendent. However, no information on other semantic
types, ontologies, nor recall is provided.
We choose MetaMap for the next experiments, as
it detects significantly larger number of concepts.
2.2 Selecting a Medical Vocabulary
This section presents the MetaMap annotation results
of eligibility criteria from 2135 breast cancer clinical
trials. The aim is to compare the coverage of criteria
by various ontologies to support the choice for further
experiments, and learn about the uncovered phrases.
Coverage by Various Ontologies. In total
MetaMap detected 768439 UMLS concepts, 10924
distinct. Figure 2 presents the statistics of their
source (left bars). Listed are only ontologies which
contributed new concepts to the set, ordered by the
number of exclusive contributions (right bars).
The majority of concepts are covered by: MTH
(UMLS Metathesaurus), CHV (Consumer Health Vo-
cabulary), NCI ( NCI Thesaurus) and SNOMED CT.
The figure demonstrates remarkable overlap between
the terminologies, emphasized by the small contribu-
tions of distinct sources (highest for NCI, SNOMED
CT and CHV). The majority of concepts (88%) are
defined by multiple ontologies. Based on the num-
ber of all detected concepts and unique contributions,
NCI seem to be the most appropriate ontology to use
for the concept recognition in eligibility criteria of
breast cancer trials. However, because SNOMED CT
is broadly used in clinical setting, and is still high on
the list, we decide to use it for the next experiments.
Uncovered Phrases. Additionally, we analyzed the
overall coverage of eligibility criteria by ontologies.
Table 2 presents the statistics about phrases distin-
guished by MetaMap. Both when using entire UMLS
and only SNOMED CT, around 32% of phrases re-
mains uncovered. To analyze the quality of obtained
HEALTHINF2013-InternationalConferenceonHealthInformatics
162
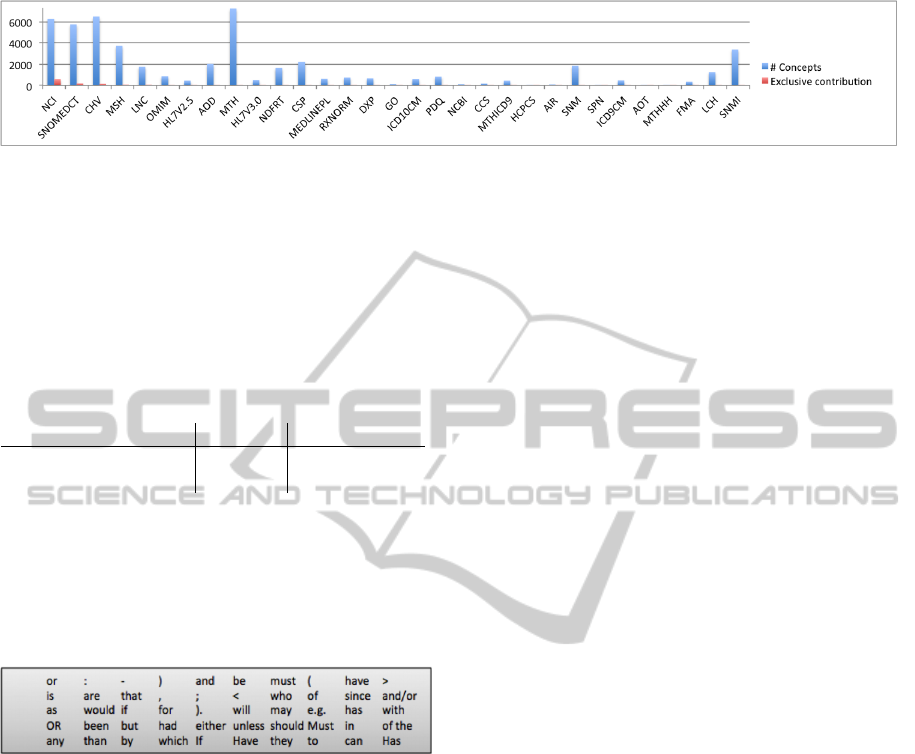
Figure 2: Number of concepts from various UMLS ontologies detected in eligibility criteria of breast cancer trials.
mappings, we checked their MetaMap score. When
using entire UMLS only 34.9% of mappings got the
maximal score, SNOMED CT, significantly more,
47.8%. UMLS is a multi-purpose source, i.e. in-
cludes concepts from vocabularies developed for dif-
ferent purposes, therefore for effective usage needs to
be customized.
Table 2: Statistics about phrases from breast cancer trials.
Phrases UMLS SNOMED CT
Uncovered 31.6 % 32 %
Max mapping sore 34.9% 47.8%
Finally, we examined the unmatched phrases (see
Figure 3), observing mainly lay terms, which is a
promising finding about UMLS coverage. However,
these provide the context, therefore their recognition
is also crucial for automated interpretation of criteria.
Figure 3: The most frequent words in eligibility criteria,
uncovered by ontologies.
3 ESTIMATING EFFORT
Using selected annotator and ontology (MetaMap and
SNOMED CT), in this section, we investigate which
parts of SNOMED CT are actually relevant for de-
scribing eligibility criteria of medical trials and esti-
mate the effort indicators for mapping terms in eligi-
bility criteria to patient data.
3.1 Distribution of SNOMED CT
Concepts over Semantic Types
Annotation of criteria with SNOMED CT resulted
in detection of 393,511 occurrences of 5994 distinct
concepts. Figure 4 presents the distribution of all de-
tected concepts over the top 25 semantic types. Fig-
ure 5 presents the cardinality of top 25 mostly repre-
sented semantic types. The most frequent concepts
have types: Qualitative Concept (13%), Temporal
Concpet (10%), Therapeutic or Preventive Procedure
(8%). The majority of distinct concepts belong to:
Disease or Syndrome (10%), Finding (7%), Organic
Chemical, Pharmacologic Substance (6%).
For mappings, the ”cardinality of the type” is an
indicator of the effort needed to map this type to pa-
tient data, while the ”frequency of the type” is an in-
dicator of how many trials will be covered by such a
mapping. In loose terms, the size of a semantic type
is the ”cost” of mapping, while the frequency is its
”benefit”. So ideally, we would like to find semantic
types with high benefit and low cost. Figure 6 shows
this benefit/cost ratio (frequency/ cardinality) corre-
sponding to the highest ranking 25 semantic types.
The situation is most ”profitable” for the type ”Re-
search activity” which occurs over 5k times and con-
tains only 4 concepts. Next are ”Patient or Disabled
Group”, ”Hormone”, ”Amino Acid, Peptide”. Only
few types contain concept that frequently occur and
are limited in number. The majority occurs sporadi-
cally with relatively large number of concepts, as the
ratio decreases very slowly. Concluding, the long tails
on the above graphs show that the mapping effort will
spread over many semantic types, and we cannot fo-
cus only on most frequent or largest types. However,
presented ordering should help to optimize the effort.
3.2 Verifying Stability of Annotation Set
By annotating the large corpus of trials we wanted to
obtain the set of concepts that is sufficiently broad to
cover the majority of trials, including those not pre-
sented in the initial corpus. To verify this idea, we an-
alyzed how the number of distinct concepts occurring
in eligibility criteria is growing with the number of tri-
als fed to the annotator. The results are plotted for the
major semantic types in Figure 7. Initially, the num-
ber of concepts grows rapidly, independently of the
type, then, the curves gradually slow down because of
the trials similarities. As expected, the number of con-
cepts belonging to some types keeps growing consid-
erably, e.g. Disease or Syndrome, while in other cases
IdentifyingMostRelevantConceptstoDescribeClinicalTrialEligibilityCriteria
163

Figure 4: Distribution of all detected concepts over semantic types.
Figure 5: Distribution of cardinality of semantic types.
Figure 6: The ratio: semantic type frequency/cardinality.
Figure 7: Growth of SNOMED concepts in eligibility crite-
ria of breast cancer trials, while increasing the trials corpus.
it stabilizes sooner, e.g. Laboratory or Test Result.
Figure 7 shows only the behavior of semantic types
with highest cardinality. The semantic types in the tail
of Figure 5 show a more promising behaviour: their
growth is small after an initial growth period, hence
for these semantic types there seem to exist a ”core
set of concepts” used in eligibility criteria. However,
we cannot expect to obtain a complete and stable set
of all concepts. Extending the trial matching system,
will require some effort of defining new mappings.
4 PRIORITIZING CONCEPTS
Performed annotation led to the recognition of sev-
eral thousands of concepts in eligibility criteria from
breast cancer trials. In this section, we prioritize them,
to suggest the order used to map the terms to pa-
tient record. Previous section provides insights ag-
gregated for semantic types. Here, we focus on con-
crete terms. To prioritize the breast cancer concepts,
apart from concept frequency we take into account
concepts specificity for breast cancer, by comparing
their usage in other trials. Next, we rank higher con-
cepts that are specific to cancer trials in general, again
as compared to their use in any trial. Furthermore, we
verify the coverage of eligibility criteria from various
trials, by the top ranking concepts in breast cancer.
4.1 Description of a Method
The concepts specificity for breast cancer was mea-
sured using the tf-idf weight (Jones, 1972), com-
monly applied in information retrieval field to detect
relevant terms (t) in a document (d) (see Formula 1).
t f ∗ id f (t, d, D) = t f (t, d) ∗ log(
|D|
|{d ∈ D}|
) (1)
HEALTHINF2013-InternationalConferenceonHealthInformatics
164

It grows proportionally to the term frequency (tf), and
inversely proportionally to the number of containing
it documents in a corpus D ( idf).
First, to rank higher concepts specific to breast
cancer, we concatenated all corresponding eligibility
criteria in one document, and as a corpus we used eli-
gibility criteria from all trials related to cancer. Anal-
ogously, to give the priority to the concepts specific
to cancer in general, we used as corpus trials studying
other diseases. We categorized the trials using their
meta data - each defines a list of studied conditions.
The numbers of applied trials are listed in table 3.
Table 3: Size of corpora used in the experiment.
Condition Trials Concepts
Breast cancer 2135 5994
Cancer 12022 13547
Non-cancer 23963 19428
4.2 Top Ranking Concepts
Using the described strategy, we obtained the order-
ing of concepts. 10 most typical for breast cancer tri-
als, and cancer in general, are listed in table 4. The
first on breast cancer list is ”Carcinoma of breast”,
cancer - ”Metastatic to”. The outcome follows the in-
tuition, demonstrating the effectiveness of tf-idf. The
obtained ranking should help to optimize the mapping
effort needed to build a recruitment support tool.
Table 4: The most relevant concepts for BC and cancer tri-
als.
Most relevant for BC Most relevant for cancer
Carcinoma of breast Metastatic to
Breast cancer Before
Invasive Chemotherapy regimen
HER-2/neu Concurrent
Concurrent
Radiotherapy
Before Chemotherapy
Specific Therapeutic procedure
Breast Malignant neoplasm
Entire breast Neoplasms - malignant
Immunologic adjuvant Radiotherapy
4.3 Coverage in other Types of Trials
Here, we present the result of the experiment aimed
to analyze the extensibility of our approach to other
diseases. We want to verify how many concepts rele-
vant for breast cancer, are also used in eligibility cri-
teria of trials studying other diseases. The trials were
clustered based on the top frequently occurring condi-
tions in the corpus of cancer and non cancer trials. We
performed the experiment with the top 2000 concepts
according to the tf-idf weight. The ordering reflects
the concepts weights of on the merged list of breast
cancer and cancer specific items. Table 5 presents the
statistics about the trials groups, overlaps of the top
breast cancer concept and the percentage of all de-
tected concepts in a group.
Table 5: Coverage of criteria related to various diseases, by
the most relevant 2000 breast cancer concepts.
Condition Trials Overlaping concepts
Prostate cancer 1214 1657 (24%)
Lung cancer 854 1662 (38%)
Lymphoma 616 1476 (42%)
Leukemia 615 1378 (42% )
Healthy 2760 1480 (21% )
HIV 1881 1430 (25%)
Obesity 844 1217 ( 31%)
Hypertension 804 1185 (34%)
The highest overlap occurs between breast and
lung cancer trials. As expected, there is a bigger over-
lap between trials about breast cancer and other can-
cers, than those about non cancer conditions (consid-
ering also the number of compared trials). In all cases
more than half of top ranking concepts for breast can-
cer, are also detected in eligibility criteria related to
other diseases. This finding indicates that the substan-
tial part of mappings can be reused if the trial match-
ing algorithm should be extended to others diseases.
5 RELATED WORK
The problem of identification of subsets of ontologies
can be compared to the problem of formal ontology
modularization. In (Clark and Parsia, 2008) the au-
thors provide an overview of existing methods, eval-
uate them from the perspective of correctness, com-
pleteness, minimality and import-safety. According
to their findings locality-based modules are proven to
be correct and complete and are empirically-shown to
approximate minimality better than ad-hoc and other
formal algorithms. These methods are applicable
when the extracted module should be sufficient for
reasoning, which is not our concern.
In (Milian et al., 2009) we aimed to detect the sub-
set of UMLS related to breast cancer treatment, by ex-
panding the initial set of concepts (those considered
at the decision points in treatment guidelines) via the
ontology hierarchy and the UMLS semantic network.
With respect to the analysis of eligibility criteria,
(Ross et al., 2010) provide an informative overview
of types of criteria, based on randomly-chosen 1000
eligibility criteria from ClinicalTrials.gov. They cate-
gorized them along several axes: complexity, seman-
tic patterns, clinical content and data sources. They
IdentifyingMostRelevantConceptstoDescribeClinicalTrialEligibilityCriteria
165

demonstrated a large semantic and clinical variability
of criteria across the trials. They argue that the ma-
jority of criteria present the challenges for automatic
evaluation because of semantic connectors hard to ex-
press with current representation languages, temporal
constraints, need for clinical judgment or lack of ex-
pected data in patient record.
6 CONCLUSIONS
The work described in this paper is part of our re-
search aimed at supporting patient recruitment and
trial study feasibility. It focuses on the analysis of se-
mantics of eligibility criteria, detecting parts of med-
ical ontologies relevant for a particular disease.
First, we investigated which annotation tool,
MetaMap or NCBO annotator, is more appropriate
for our task. We compared the overlap of concepts
detected by both in eligibility criteria of 2135 breast
cancer trials. The results show that the intersection
accounts for only 59% of entire set. Because of the
advantage of MetaMap in the number of detected con-
cepts we decided to use it for further experiments. In
future work it could be interesting to define a voting
algorithm which takes into account precision and re-
call of both tools corresponding to particular types
of criteria or semantic types. Second, we analyzed
the source and semantic types of detected concepts.
The findings indicate the high majority of concepts
(88%) is defined by more than one ontology cov-
ered by UMLS, majority by MTH, CHV, NCI and
SNOMED CT. The highest number of unique contri-
butions is provided by NCI, SNOMED CT and CHV.
We chose SNOMED CT for the next experiments, be-
cause of its wide usage in clinical setting and good
scores in the comparison. It should be noted that in
32% of criteria phrases MetaMap did not detect any
concept, which indicates that additional processing is
needed to recognize the context in which recognized
terms occur. Only approximately 35% of phrases an-
notated with UMLS obtained the maximal mapping
score, and 48% in case of using only SNOMED CT.
The analysis of the distribution of the detected
concepts over various semantic types and their fre-
quency revealed that the mapping effort will need to
be spread over many types. Furthermore, we analyzed
the stability of obtained concept set by studying its
growth while adding new trials. While some stabil-
ity of the growth curve can be observed, specially for
some semantic types, we cannot expect that obtained
annotation set is complete. Extending the solution to
other trials will involve creating more mappings.
Finally, we put the semantic of breast cancer trials
into broader perspective of over 38, 000 clinical trials
studying other diseases. We used tf-idf measure to
find concepts that are specific for breast cancer, and
cancer in general, and used the results to prioritize
them. We also verified the overlap between the top
2000 ranking concepts for breast cancer and concepts
occurring in other types of eligibility criteria and find
out that the substantial part is repeated: in all cases
above 1100, in other cancer types above 1300.
We believe that this analysis provides insights
about semantics of eligibility criteria that can be used
to prioritize the mapping process of eligibility crite-
ria to patient record, and enhance building the re-
cruitment support tool. The approach was demon-
strated on the breast cancer domain, but it can be eas-
ily reused for other diseases.
REFERENCES
Aronson, A. R. and Lang, F.-M. (2010). An overview of
metamap: historical perspective and recent advances.
Journal of the American Medical Informatics Associ-
ation, 17(3):229–236.
Clark, K. and Parsia, B. (2008). Modularity and owl. Liter-
ature survey.
Jones, K. S. (1972). A statistical interpretation of term
specificity and its application in retrieval. Journal of
Documentation, 28:11–21.
Milian, K., Aleksovski, Z., Vdovjak, R., ten Teije, A.,
and van Harmelen, F. (2009). Identifying disease-
centric subdomains in very large medical ontolo-
gies, a case-study on breast-cancer concepts in
snomed. In Knowledge Representation for Healthcare
(KR4HC09), LNCS.
Milian, K., Bucur, A., and ten Teije, A. (2012). Formaliza-
tion of clinical trial eligibility criteria: Evaluation of a
pattern-based approach. In 2012 IEEE International
Conference on Bioinformatics and Biomedicine.
Musen, M., Shah, N., Noy, N., Dai, B., Dorf, M., Griffith,
N. B., Buntrock, J., Jonquet, C., Montegut, M., and
Rubin, D. (2008). Bioportal: Ontologies and data re-
sources with the click of a mouse. In AMIA Annual
Symposium, pages 1223–1224.
Ross, J., Tu, S. W., Carini, S., and Sim, I. (2010). Anal-
ysis of eligibility criteria complexity in clinical trials.
AMIA Summits on Translational Science Proceedings,
pages 46–50.
Shah, N. H., Bhatia, N., Jonquet, C., Rubin, D. L., Chi-
ang, A. P., and Musen, M. A. (2009). Comparison of
concept recognizers for building the open biomedical
annotator. BMC Bioinformatics, 10(S-9):14.
HEALTHINF2013-InternationalConferenceonHealthInformatics
166
