
StorageBIT
A Metadata-aware, Extensible, Semantic and Hierarchical Database for Biosignals
Carlos Carreiras
1
, Hugo Silva
1
, Andr
´
e Lourenc¸o
1,2
and Ana Fred
1
1
Instituto de Telecomunicac¸
˜
oes, IST-UTL, Lisbon, Portugal
2
Instituto Superior de Engenharia de Lisboa, ISEL, IPL, Lisbon, Portugal
Keywords:
Biosignal, Database, Metadata, File Format, Semantics.
Abstract:
Acquisition of biomedical data is, nowadays, widespread, originating a deluge of data that may contain rele-
vant and interesting information for health-care professionals, biosignal researchers, and the individuals them-
selves. This creates the need to organize the information in a structured way, facilitating collaboration and
research efforts. Therefore, for that purpose, this paper investigates database systems and file formats, dis-
cussing current technologies, requirements and possible implementations. These implementations were put
through a benchmarking package to analyze their insertion, query and update performance. A final approach
combining the use of HDF5, a hierarchical file format for numerical data, and MongoDB, a NoSQL database,
is proposed, as it showed the best combination of properties from the tested solutions.
1 INTRODUCTION
The ability to measure biomedical signals (biosignals)
is no longer restricted to specialized environments
and expensive equipment. Indeed, recent advances
in wireless protocols, ubiquitous computing, and sen-
sor technology have made the acquisition of biosig-
nals widespread. Companies like Nike
1
, Zeo
2
, and
Fitbit
3
all currently commercialize products to track
various types of information, such as the number of
steps, calories, sleep, weight, heart rate, etc.. This
technological revolution has given way to a new con-
cept of healthy living, the so-called Quantified Self
movement, whereby objective, quantified information
can lead to insights that prompt behavior change, re-
sulting in improved health (Kvedar, 2011).
This opens access to vast amounts of data that,
first and foremost, need to be stored. Only then
is it possible to apply signal processing algorithms
(Vaseghi, 2006), pattern recognition, multi-modal
data fusion and classification techniques (Jain et al.,
2000) to extract meaningful information from the ac-
quired signals. This work is thus focused on the prob-
lem of data storage and access in the context of biosig-
nals, exploring the requirements, available technolo-
gies and efficiency aspects of choosing a database sys-
1
www.nike.com/fuelband
2
www.myzeo.com/sleep
3
www.fitbit.com
tem and developing its interface. This is done in two
perspectives. First, the format on which the data is ac-
tually stored must be specified and implemented, and
second, the database itself needs to be deployed.
The remainder of this paper is organized as fol-
lows. We start with a review of the state-of-the-
art, pointing out limitations of current approaches.
Then, the basic requirements are presented, dis-
cussing available technologies for the storage of
biosignals. Subsequently, various implementation al-
ternatives are analyzed, and then evaluated with a set
of performance tests. Finally, the main conclusions
are outlined, specifying the chosen implementation
and future work.
2 STATE-OF-THE-ART
Databases for biosignals are essential for scientific
development, research in biomedical signal process-
ing and in health-care applications. However, exist-
ing attempts to standardize a file format for the stor-
age and sharing of biosignals have several limitations,
which hamper research efforts. Indeed, standardiza-
tion allows for easier collaboration, which improves
the quality of developed software, exchange of sci-
entific results and health-care practices, while also
reducing costs, and leading to the discovery of new
knowledge (Varri et al., 2001). In reality, many file
65
Carreiras C., Silva H., Lourenço A. and Fred A..
StorageBIT - A Metadata-aware, Extensible, Semantic and Hierarchical Database for Biosignals.
DOI: 10.5220/0004241400650074
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2013), pages 65-74
ISBN: 978-989-8565-37-2
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

formats exist, with well over 100 documented for-
mats (Brooks et al., 2011), such as the Extensible
Biosignal File Format (EBS) (Hellmann et al., 1996),
the European Data Format (EDF+) (Kemp and Oli-
van, 2003), the Medical Waveform Format Encod-
ing Rules (MFER) (MFER, 2003), and the WaveForm
DataBase (WFDB) (Goldberger et al., 2000). Many
formats are proprietary, limiting the access to the in-
formation contained in the files. Most are designed
for a specific purpose, such as the storage of electro-
cardiogram (ECG) signals, and there is a general lack
of structured metadata, that is, all the semantic infor-
mation that describes how the signals were acquired
and what was done with them.
It should be noted that peer-reviewed literature on
this topic is fairly limited. A quick search for ”biosig-
nal database” in IEEE’s Xplore
R
Digital Library pro-
duces 28 results. Of these, only four directly address
the issues of specifying and implementing a system
for the storage of biosignals. For instance, Penzel et
al. (Penzel et al., 2001) describe the approach used
to store polysomnography (PSG) data, produced in
the scope of SIESTA, an European project carried out
in 2001. The European Data Format was used, with
the need to specify strict filename conventions (e.g.
encoding the subject identification, or the recording
site), and the internal structure of the files (i.e. the or-
der of the signals in the file and their sampling rates).
In the same year, Lovell et al. (Lovell et al., 2001) de-
tailed a framework for web-enabled storage of biosig-
nals, where it is already noted the absence of a com-
mon file format. The development of browser-based
applications is discussed and access-load issues are
addressed.
More recently, the focus has been given to se-
mantic approaches (see, for example, (Brooks et al.,
2011), (Kokkinaki et al., 2008), and (Brooks, 2009)),
where, in addition to the core biosignal data, contex-
tual information is included, such as patient and ac-
quisition procedure information, enabling the integra-
tion of disparate and heterogeneous sources of med-
ical information and facilitating their query and re-
trieval.
It is easy to grasp that current methods exhibit
some glaring limitations. In particular, it is common
to have the signal data separated from metadata char-
acterizing the experimental setup. Therefore, possess-
ing the data files alone is of limited interest, because
the acquisition context necessary to analyze them can-
not be accessed. Furthermore, current file formats
usually have a fixed number of metadata fields, with
limited size, employing weak semantics (while cer-
tain metadata fields are self-explanatory – sampling
frequency, resolution, etc. – others may not be – e.g.
”channel” may refer to the actual samples of a sig-
nal, its label, or the identification of the physical in-
put of the acquisition system). Additionally, current
approaches make it hard to append new information
to an already existing file (e.g. a filtered version of
the biosignals, or a comment about the data). And,
finally, these approaches provide poor interfaces to
the user, with very limited or inexistent query sup-
port. These limitations provide our motivation to
build upon the current state-of-the-art and develop an
extensible, semantic and hierarchical infrastructure.
3 STORING BIOSIGNALS
In response to the limitations found in previous work,
a list was curated with the most important aspects
and properties a system that stores biosignals should
exhibit. Based on these requirements, a data model
was specified and various implementations of the data
model were investigated.
3.1 Requirements
The following properties were used to evaluate and
compare the various file formats under study: 1) Ac-
cess Performance: Read and write speed, non-
sequential access, data compression, etc.; 2) Cross-
Platform Support: The availability of tools for
different operating systems and programming lan-
guages; 3) Events Support: Events and annotations
are textual comments or values related to a particular
signal, or to the acquisition session as a whole (e.g.
the location of R waves in the ECG). This type of data
is very important, given that only through the evalua-
tion of annotations a human user or a computer algo-
rithm can learn the meaning of specific signal patterns
(Penzel et al., 2001); 4) Extensibility: The ability to
easily add more data to a file; 5) Metadata: Defined
as data about data, pertains to all the additional infor-
mation that characterizes the acquired signals. It in-
cludes general and particular attributes of the biosig-
nals, when, how and by whom the acquisitions were
made, their purpose, and what processing has been
applied. Fields for metadata should be extensible (al-
lowing to add more information along the way) and,
more importantly, should have meaning, this is, the
use of a controlled vocabulary to specify content, e.g
using ontologies (McGuinness, 2003). Some com-
mon metadata fields can be seen in Figure 1. The
use of metadata allows for knowledge to be processed
computationally in a comparable way to numeric data
(Brooks, 2009); 6) Multi-modality: The capability to
store various signal types in a single container struc-
HEALTHINF2013-InternationalConferenceonHealthInformatics
66

Table 1: Comparison of file formats and NoSQL DBMSs.
(a) Comparison of file formats.
Property
File Format
EBS EDF+ MFER WFDB HDF5
Compression - - - - +++
Non-Sequential Access - - - - +++
Cross-Platform Support + ++ ? ++ ++
Events Support ++ + - ++ ++
Extensibility - - - - +++
Metadata ++ + + + +++
Multi-modality - + - + +++
Querying - - - - -
(b) Symbols used in Table (a).
Symbol Description
? Information Not Available
- Not Supported
+ Supported
++ Good Support
+++ Excellent Support
(c) Comparison of NoSQL database management systems.
Property
NoSQL DBMS
CouchDB MongoDB
Flag Line
DB consistency, ease of use
Scalable, high-performance, retains
friendly properties of SQL
Interface
HTTP/RESTful BSON
Queries
MapReduce Javascript expressions
Replication
Master-Master Master-Slave, replica sets
Others
Multiversion Concurrency Control,
attachment handling
Memory-mapped files, sharding,
embedded file system (GridFS)
Rec o rd = {
’ Header ’: { ’ Sess i o n ’: ’#’ ,
’ Date ’: ’ yyyy -mm - d d Thh : mm : ss ’ ,
’ Sup e r viso r ’: ’ txt ’ ,
’ Exp e r imen t ’: { ’ Name ’: ’ txt ’ ,
’ Des c rip t ion ’: ’ txt ’,
’ G oals ’: ’ txt ’} ,
’ Subject ’: { ’ Name ’: ’ txt ’ ,
’ Birthda y ’: ’ yyyy - mm - dd ’,
’ Ge n d e r ’: ’txt ’ ,
’ Contact ’: ’ txt ’}} ,
’ Tags ’: [ ’ txt ’, ...] ,
’ A udit ’: {} ,
’ Bios i g nal ’: { ’ Type ’: ’ txt ’,
’ De v i c e ’: ’txt ’ ,
’ Sa m ple Rate ’: ’#’ ,
’ Duratio n ’: ’# ’ ,
’ Res o l utio n ’: ’# ’ ,
’ U nits ’: { ’ Time ’: ’ txt ’ , ’ Sensor ’:
’txt ’}} ,
’ Ev e n t s ’: { ’ Source ’: ’ txt ’ ,
’ Type ’: ’ txt ’ ,
’ Dic t i onar y ’: { ’ key ’: ’ value ’}} }
Figure 1: Hierarchical structure of a Record, in a JSON-style notation, as specified by the Data Model, with some common
metadata fields.
ture. These signals may be of different nature (e.g.
ECG, EEG, PSG, etc.), and possibly acquired at dif-
ferent sampling rates, or even be mere events, thus
requiring appropriate accompanying information and
data structures; and 7) Querying: A query is a precise
request for retrieval of information that shares com-
mon properties within a database or an information
system. In particular, we are interested in the inclu-
sion of a system of tags, generated by biosignal re-
searchers and experts, leveraging the potential power
of folksonomies (Wal, 2007). In Table 1(a) the EBS,
EDF+, MFER and WFDB file formats are compared
based on these criteria.
Additionally to these requirements, the three fol-
lowing general aspects should also be taken into con-
sideration: 1) Accessibility: Data must be accessi-
ble to multiple people, possibly in multiple locations.
Therefore, the information must live on the cloud,
taking advantage of current web-based technologies;
2) Scalability: The ability of a system to respond to a
growing amount of work. This can be done in a verti-
cal sense (by adding more resources to a single node
in a system) or in a horizontal sense (by adding more
nodes to a system). Additionally, the intrinsic soft-
ware elements of the system should also be as efficient
as possible. In the context of biosignals, the bottle-
neck lies, predominantly, in storage constraints. For
instance, a night-long acquisition of PSG data easily
exceeds the 1GB threshold. Thus, the system must
deal with large data volumes, and must process it (i.e.
put and retrieve operations) quickly; and 3) Flexibil-
ity: The system must sort through records from dif-
ferent experiments and through data of various types.
Therefore, it needs to be flexible enough to accommo-
date these different needs, and to be able to cope with
changing requirements.
StorageBIT-AMetadata-aware,Extensible,SemanticandHierarchicalDatabaseforBiosignals
67

Start Date Start Time
Duration
Version
Reserved
Number
of Bytes
Number of
Data Records
Recording ID
Patient ID
( NS xLabel
Transducer
Physical Dimension
Physical Maximum
Digital Maximum
Digital Minimum
Preltering
Number of samples
Reserved
( NS x
( NS x
( NS x
( NS x )
)
)
)
)
)
)
)
)
( NS x
( NS x
( NS x
( NS x
Physical Minimum
( NS x )
HEADER
NS
(a) EDF+ header structure, with fixed-length fields on the left and variable-length fields on the right, where each sharp-edged square represents 8 ascii
characters (64 bytes).
Signal 1 )
( Samples x
. . .
Signal NS )
( Samples x
DATA RECORD
(b) EDF+ data record structure, storing multiple (NS) biosignals; each round-edged square represents one byte.
Figure 2: Schematic representation of the EDF+ file structure; NS is the number of signals stored in the file.
3.2 Data Model
The basic abstract entities for storage are Records.
Records are equivalent to acquisition sessions, in the
sense that all the signals from one acquisition session
are stored in one Record. The signals should be con-
textualized with the experiment (i.e. the purpose of
the acquisition), the details of the subject, and com-
plemented with any relevant metadata, such as a de-
scription of the processing steps necessary to obtain
the signal (to audit the processing method), the phys-
ical units of the signals, the labels of the channels, or
annotations made by the acquisition’s supervisor. In
this way, multiple users could access the same data,
process it, and share any obtained results in a way
that is understandable by everyone, in a collaborative
effort.
With these principles in mind, an integrated
database management system for biosignals was de-
signed, in which the acquired records are sorted
by subject and, subsequently, by experiment. The
Records are composed by four main fields (see Fig-
ure 1): 1) Header: Identifies the file and contains
basic information about the stored biosignals, spec-
ifying date and time of acquisition, who performed
the acquisition to whom, the experimental context,
and any automatically or manually generated index-
ing tags; 2) Audit: This field is intended to list a de-
tailed history of the file, i.e., a systematic review of
what was done to the biosignals. It would summa-
rize, for instance, which filters were applied, together
with the corresponding parameters, or the processing
steps used; 3) Biosignals: The signals of interest, i.e.,
synchronous data; 4) Events: Any events or annota-
tions made asynchronously from the recorded biosig-
nal time series.
It should be noted that no element of the data
model has a predefined or expected volume of infor-
mation, being as big as necessary and extensible. Fur-
thermore, the model is designed to be open. Elements
can be added or removed, according to the use case.
This makes it possible to adapt the model to any type
of field structure, and, therefore, current file formats
can be easily mirrored in the Data Model.
As an example of how the Data Model compares
to one of the most widely used biosignal file formats,
the internal structure of the EDF+ format is shown in
Figure 2. An EDF+ data file is composed by a header
followed by data records. The header identifies the
subject and describes the technical parameters of the
recording, while the data records contain the time se-
ries data. Although the header has variable-length, it
HEALTHINF2013-InternationalConferenceonHealthInformatics
68

is only to support the multiple data records stored in
the file, as the fields for each data record themselves
are dimensionally limited. Furthermore, the format
does not allow to store additional, user-defined meta-
data fields. Figure 3 shows how the proposed Data
Model can be used to mirror the EDF+ format. The
main difference is that, with the Data Model, meta-
data pertaining to a certain biosignal is stored in the
same hierarchical node as that biosignal, thus explic-
itly defining the ownership of the metadata. Addition-
ally, it is easy, with this approach, to retrieve just one
of the stored biosignals, without being necessary to
parse the entire file. This is done by directly navi-
gating to the desired hierarchical node, where all the
necessary information is stored. Note that the Data
Model requires the definition of the metadata fields
for each stored biosignal. This implies some over-
head, both in computational terms and to the user,
but it is our opinion that the flexibility and more
detailed information this structure provides compen-
sates the additional workload. As stated earlier, the
Data Model encourages the definition of the metadata
fields according to each application case, and, accord-
ingly, the biosignal nodes may contain varying sets of
metadata fields.
3.3 Implementation
Having described the abstract data model, it is now
relevant to discuss how to put it into practice. To that
end, the two technologies presented below, NoSQL
databases and the Hierarchical Data Format, were
considered since they exhibit a set of properties that
meet the previously discussed requirements.
3.3.1 NoSQL Databases
As the name suggests, NoSQL is a family of Database
Management Systems (DBMS) that does not use the
SQL language, and many times does not employ the
relational model at all (Strauch, 2011). These stor-
age systems do not need fixed schemas, and typically
scale horizontally. They are optimized for retrieve and
append operations, and are capable of dealing with
large amounts of data, making them ideal to solve the
problem at hand. Typical NoSQL categories include
key-value stores, Big Table implementations, docu-
ment stores and graph databases (Strauch, 2011). Of
these, the most interesting in the context of biosignals
is the document-based approach.
The basic notion of this type of DBMS is that
a document encapsulates and encodes data in some
standard format (e.g. XML, JSON). Documents are
analogous to rows in the SQL language, but they do
not need to comply all with the same schema, i.e.,
Rec o rd = {
’ He a d e r ’: {
’ Subject ’: < P ati ent ID >
’ Re c ord ID ’: < Re c ord ID >
’ Date ’: < S tart Date > + < S t art
Time >
} ,
’ Bi os i gn a l 1 ’: {
’ Duratio n ’
’ Sa m ple Rate ’
’ L abel ’: < L a b el [1] >
’ Tra n s duce r ’: < T ra n sd uc e r [1] >
’ Ph ysi cal Di m e nsion ’: < P h ys i ca l
Di m en s io n [1] >
’ Ph ysi cal Maximum ’: < Ph ysi cal
Ma x imu m [1] >
’ Ph ysi cal Minimum ’: < Ph ysi cal
Mi n imu m [1] >
’ Di g it a l Max i m u m ’: < D i gi t al
Ma x imu m [1] >
’ Di g it a l Min i m u m ’: < D i gi t al
Mi n imu m [1] >
’ A udit ’: < P re fi l te ri ng [1] >
} ,
...
’ Bi os i gn a l NS ’: {
’ Duratio n ’
’ Sa m ple Rate ’
’ L abel ’: < L a b el [ NS ] >
’ Tra n s duce r ’: < T ra n sd uc e r [ NS ] >
’ Ph ysi cal Di m e nsion ’: < P h ys i ca l
Di m en s io n [ NS ] >
’ Ph ysi cal Maximum ’: < Ph ysi cal
Ma x imu m [ NS ] >
’ Ph ysi cal Minimum ’: < Ph ysi cal
Mi n imu m [ NS ] >
’ Di g it a l Max i m u m ’: < D i gi t al
Ma x imu m [ NS >
’ Di g it a l Min i m u m ’: < D i gi t al
Mi n imu m [ NS ] >
’ A udit ’: < P re fi l te ri ng [ NS ] >
}
}
Figure 3: Mirroring of the EDF+ file structure onto the Data
Model, using a JSON-style notation; text between ”<>”
identifies the corresponding field in the EDF+ format.
they do not need to possess the same fields, which
fits nicely the previously mentioned requirements. In
this work, the CouchDB (Anderson et al., 2010) and
MongoDB (Chodorow and Dirolf, 2010) implemen-
tations were analyzed. The reason for choosing these
two document-based NoSQL DBMSs resides in the
fact that both encode the data in JSON (BSON – bi-
nary JSON – for MongoDB), which is becoming in-
creasingly popular as a language-independent, data-
interchange format (Crockford, 2006). Table 1(c) pro-
vides a comparison of MongoDB and CouchDB.
StorageBIT-AMetadata-aware,Extensible,SemanticandHierarchicalDatabaseforBiosignals
69

Table 2: Implementation alternatives for the desired biosignal storage system; solution ”A4” is the proposed approach.
Type Description Code Technologies
DB Only
All data and metadata stored in the
DB
A1.1 MongoDB
A1.2 CouchDB
File Only
All data and metadata stored in
HDF5 files
A2 HDF5
File in DB
All data and metadata stored in the
DB, but data files are attached to
documents on the DB
A3.1
MongoDB,
HDF5 via
mongofiles
A3.2
MongoDB,
HDF5 via
rawIO
File with
DB
Metadata stored in DB, data stored
in files served out of a dedicated
and distributed file system
A4
MongoDB,
HDF5
3.3.2 Hierarchical Data Format (HDF5)
The Hierarchical Data Format is a self-describing data
format designed to store and organize large amounts
of numerical data (HDF, 2010). It is supported by
many software platforms, including C/C++, Java,
Matlab and Python. The file structure is based on two
major types of objects: Datasets (multidimensional
arrays of a homogeneous type), and Groups (con-
tainer structures which can hold datasets and other
groups). This model is very similar to the philosophy
behind the design of JSON files, being completely
flexible, and thus allowing the storage of annotations
and all the desired metadata. In particular, it is possi-
ble to define attributes for both the Dataset and Group
objects, storing the metadata associated with them.
Datasets can be compressed (GZIP), and can also be
extended and accessed non-sequentially, an advantage
resulting from the use of B-Trees (Bayer, 1971). It is
also possible to link to other HDF5 files. It is obvious
from this discussion, and from the comparison made
in Table 1(a), that the HDF5 file format is the one that
best covers the listed requirements, with the exception
of the querying property. Therefore, a simple query
engine was implemented to search the HDF5 files.
3.4 Implementation Alternatives
Making use of MongoDB, CouchDB and HDF5, we
implemented and evaluated the performance of the
following approaches: 1) DB Only: All data and
metadata is stored in the DB; this was done using
MongoDB (case A1.1) and CouchDB (case A1.2);
2) File Only: All data and metadata is stored in
HDF5 files (case A2); 3) File in DB: All data and
metadata is stored in the DB, but HDF5 files with
the signal data are attached to documents on the DB;
using MongoDB, two alternatives for file attachment
were tested, one using mongofiles
4
(case A3.1), the
other using rawIO
5
(case A3.2); and 4) File with DB:
Metadata stored in MongoDB, data stored in HDF5
files served out of a dedicated and distributed file sys-
tem (case A4). Solution A4 is the proposed approach.
This information is summarized in Table 2.
4 PERFORMANCE RESULTS
The task of evaluating a DB system is not an easy one.
It should be noted that there is no single evaluation
measure that universally classifies a DB system, as
it is highly dependent on the specific application. In
this sense, the first question that should be posed is the
question of applicability, i.e., if the system solves the
problem at hand. Secondly, the system is to be used
by people from different backgrounds and, as such, it
should be easy to use. Finally, performance measures
like speed, persistency and scalability should also be
addressed.
Taking into account the various approaches de-
scribed above, several tests were designed for exper-
imental evaluation, in order to assess their perfor-
mance. In particular, insertion, update and query op-
erations were measured, and in order to have a real-
istic test environment, a simulation setting was cre-
ated. This base setting is composed of 100 experi-
ments, 1000 subjects and 1000 records (1 per subject,
10 per experiment). For each record, a dummy biosig-
nal (all zeros) was added, with 32 channels, lasting for
900 seconds and sampled at 1024 Hz. Each element
of the signal is a 16-bit integer, which accounts for a
4
A command line tool included in the MongoDB distri-
bution to access GridFS, its embedded file system.
5
An alternative to the mongofiles tool using the Python
interface to handle data streams.
HEALTHINF2013-InternationalConferenceonHealthInformatics
70

Figure 4: Results of the Biosignal Storage test.
total size of 56.25 MB per record, or about 55GB for
the entire base set. On top of this base set, the follow-
ing performance tests were applied: a) Insertion of
metadata fields; b) Biosignal Storage; c) Biosignal
Retrieval; d) Query of the metadata fields; e) Up-
date in Place of metadata fields, without changing
the size of the field; and f) Add Update of metadata
fields, adding a new field.
The metadata associated with each record is com-
posed by the names of the experiment and the subject,
and a date string (ISO8601 format). The metadata
associated with each dummy biosignal comprises the
sample rate, resolution, duration, a type (i.e. ’zeros’),
and a list of channel labels.
The tests were applied in a cumulative manner,
i.e., by first adding a number of new elements to the
base set, and then executing the tests. This was re-
peated for 10, 50, 100, 500 and 1000 new additions,
so that the final set had 1760 experiments, 2660 sub-
jects and 2660 records, with a total size of about
170GB, taking into account all signal data and meta-
data. The performance (P) of the Insertion, Query,
Update in Place and Add Update tests was measured
in operations per second (ops), by timing how long
each operation took to run (t
i
), and then dividing the
number of operations (N) by the total time (Equation
1):
P =
N
∑
N
i=1
t
i
(1)
The rate (R) of the Biosignal Storage and Retrieval
operations was quantified in MB/s by measuring the
amount of data that was being added or retrieved in
each operation (d
i
), timing that operation (t
i
) and then
computing a mean data rate (Equation 2):
R =
∑
N
i=1
d
i
∑
N
i=1
t
i
(2)
The tests were run on an Ubuntu machine (v10.10,
64 bits, Intel i7 with 12 cores at 3.33GHz, 16 GB
of RAM), with the necessary code implemented in
Python (v2.6.6). Version 1.0.1 of CouchDB, version
2.0.2 of MongoDB and version 1.8.4 of HDF5 were
used. All elements were run locally in order to reduce
the variability introduced by network load.
Biosignal Storage and Retrieval rates for the var-
ious cases are shown in Figures 4 and 5. In the
case of Biosignal Storage (Figure 4), the fastest re-
sults were obtained for approaches A” and A4, with
a data rate consistently above 100MB/s. A similar
result for these two approaches was already expected,
as the underlying storage technology (HDF5) is com-
mon to them. The less performing results correspond
to solution A1.2. Figure 5 shows the results for the
Biosignal Retrieval tests. Again, the approaches A2
and A4 produce the highest reading rates, although
only while the number of signals is not too big. After
1100 records for A4 and after 1500 records for A2, all
the alternatives perform similarly, at about 100MB/s.
This reduction in performance is possibly due to the
saturation of the system buffers, as the first operations
are extremely fast.
Results (mean and standard deviation) for the In-
sertion, Update and Query performance tests are sum-
marized in Table 3, where it is possible to observe
that the approaches using MongoDB (including the
proposed approach) outperform the other solutions
in every case, except for the Record Header Inser-
StorageBIT-AMetadata-aware,Extensible,SemanticandHierarchicalDatabaseforBiosignals
71
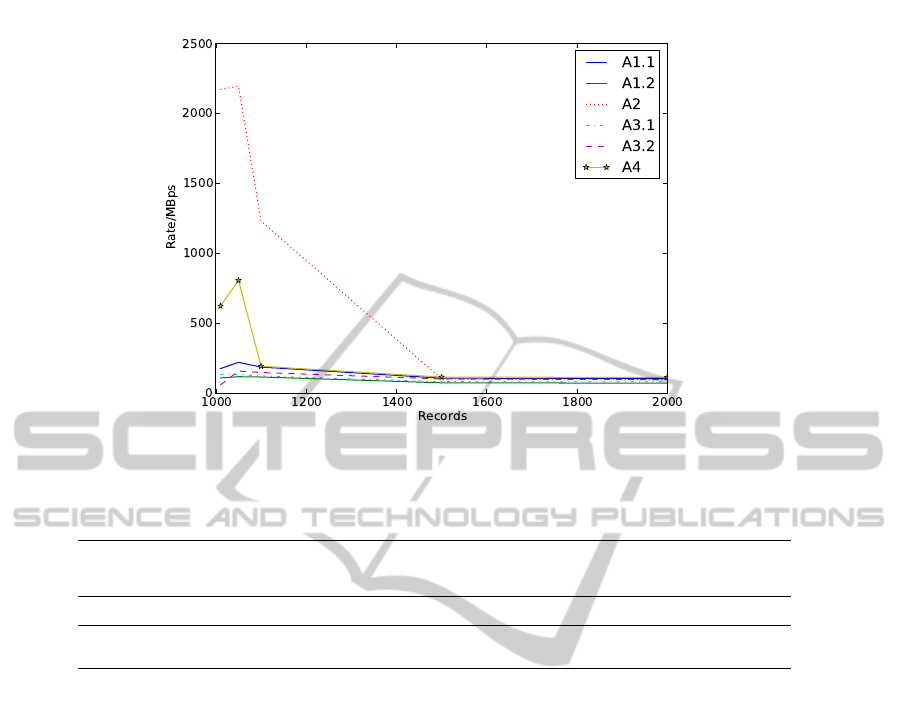
Figure 5: Results of the Biosignal Retrieval test.
Table 3: Results (µ ± σ) of the Insertion, Update and Query tests (in operations per second).
Test
Approach
A1.1 A1.2 A2 A3.1 A3.2 A4
Insertion
experiments 962.4 ± 173.7 14.1 ± 2.7 NA 930.7 ± 113.1 1086.9 ± 290.8 1002.5 ± 210.1
subjects 650.7 ± 360.0 15.6 ± 0.3 NA 821.2 ± 287.2 814.8 ± 417.3 812.9 ± 531.5
record header 220.3 ± 81.0 3.7 ± 0.02 930.1 ± 175.6 231.4 ± 103.5 289.3 ± 44.9 175.2 ± 59.6
Query
889.4 ± 50.6 5.3 ± 3.0 0.4 ± 0.2 849.0 ± 25.0 1100.1 ± 171.4 1097.7 ± 403.8
Update
in place 13100.4 ± 4966.7 19.1 ± 0.4 2235.8 ± 1061.7 10930.1 ± 6448.6 14809.4 ± 4383.2 660.3 ± 359.9
add 14346.8 ± 2973.6 19.0 ± 0.6 2797.3 ± 31.5 12907.9 ± 6205.4 17096.7 ± 4238.1 818.7 ± 35.4
tion in case A2. Note that, in this table, there are
no Experiments or Subjects Insertion results for so-
lution A2 given this solution is solely based on HDF5
files, where the creation of a new Record already in-
cludes all the metadata. Solution A1.2, employing
CouchDB, is the least performing one, in particular
for the Querying test. Indeed, there is not, at the
moment, a straightforward method the make general
queries (e.g. retrieve all documents with a specific
field matching an input value) in CouchDB. Instead,
it uses views (in a MapReduce philosophy (Dean
and Ghemawat, 2004)), which list the documents that
match a certain criterion, but these views must be de-
fined a priori, therefore limiting the scope of what is
queryable to the existing set of views. Additionally,
the interactive creation of views is not feasible, be-
cause this process is expensive to compute, as a new
index must be built, thus impacting the overall per-
formance of the system. On the other hand, Mon-
goDB’s query engine, based on memory-mapped files
and learning from the stored data, is highly efficient,
and consequently provides superior performance, re-
gardless of the existence of an index. Comparing the
MongoDB solutions (A1.1, A3.1, A3.2 and A4), the
results are similar, taking into consideration the stan-
dard deviations, except for the Record Header Inser-
tion and Update tests, where approach A4 is slower,
as a result of having to insert or update the metadata
both in the MongoDB database and in the HDF5 file.
Regarding the observed errors for the Update tests,
note that, for example in case A3.2, the mean oper-
ation time is lower than 7 × 10
−5
seconds. This is a
quite small value to measure and, therefore, any small
imprecisions lead to big errors in the resulting met-
ric, which, by Equation 1, depends on the inverse of
the measured time. Overall, we believe that solution
A4 provides the best combination of properties, uni-
fying the efficient storage and retrieval of biosignal
data provided by HDF5 files, with the fast and flexible
Insert, Query and Update operations of MongoDB.
5 DISCUSSION AND
CONCLUSIONS
In this work, the topic of storing and accessing biosig-
nals was addressed in a metadata-aware, extensible,
semantic and hierarchical way. In particular, a data
HEALTHINF2013-InternationalConferenceonHealthInformatics
72

model was defined and evaluated, database technolo-
gies and file formats were presented, and various im-
plementations were evaluated using a series of bench-
marking tests for biosignal storage and retrieval, data
insertion, update and querying. The results show
that, first of all, MongoDB is, generally, faster than
CouchDB as a DBMS. This is due to the fact that
MongoDB uses memory-mapped files, keeping the
more frequently accessed information in Random Ac-
cess Memory (RAM). Regarding the operations of in-
serting and retrieving biosignal data, the most effi-
cient approach is using the HDF5 file format. Based
on these results, our work proposes a hybrid solu-
tion (solution A4), which uses MongoDB as a front-
end for data access (stores metadata), and HDF5 as
the backbone for data persistency (stores synchronous
time series data and asynchronous event-based data),
as can be seen in Figure 6. In this way, metadata is
put under the spotlight, as it is by querying the meta-
data that a user of the system has access to the desired
signals, stored in an HDF5 file. Therefore, the pro-
posed system combines fast querying, insertion and
retrieval of biosignal data that is easily accessed from
a variety of systems and platforms. Using HDF5 files
also has the additional advantage of enabling the shar-
ing of individual records by conventional (file-based)
methods.
SERVER
CLIENT
MONGO DB
StorageBIT API
MERGER
UNISON
UNISON
HDF5 STORAGE
.EXP A
.EXP B
.EXP C
HDF5 STORAGE
.EXP A
.EXP C
SYNC OF
HDF5 FILES
QUERY
Figure 6: Schematic structure of the StorageBIT system.
Figure 6 details the overall architecture of the sys-
tem, where a centralized machine hosts the MongoDB
server and the HDF5 storage. On the client side, the
user has access to the information by sending query
requests to the server, and a local storage of the de-
sired HDF5 files (organized by experiment) is main-
tained, improving speed of access. The HDF5 storage
on the client is kept in sync with the server with Uni-
son (Pierce and Vouillon, 2004), which automatically
propagates changes from one replica to the other. The
StorageBIT module is part of an integrated biosig-
nal acquisition, storage, visualization, annotation and
processing framework, the Biosignal Igniter Toolkit
(BIT), which is currently under development at out
research group.
The proposed system thus extends the state-of-
the-art in the field, overcoming many of the common
problems faced by biosignal researchers, such as han-
dling the metadata, no size limits to header and meta-
data information, and seamless distributed access to
the data repositories. Due to the underlying data per-
sistency technology, our system is fully compatible
with existing standards, as the Data Model can be
shaped to mirror any commonly used field structure.
Further work includes the specification of the for-
mat for the Audit field, which is intended to function
as a detailed history of the various processing steps
applied to the stored signals.
ACKNOWLEDGEMENTS
This work was partially funded by Fundac¸
˜
ao
para a Ci
ˆ
encia e Tecnologia (FCT) under
grants SFRH/BD/65248/2009, SFRH/PRO-
TEC/49512/2009, PTDC/EIA-CCO/103230/2008
and PTDC/DES/103178/2008. The authors would
also like to thank the Institute for Systems and
Technologies of Information, Control and Communi-
cation (INSTICC), the graphic designer Andr
´
e Lista,
Prof. Pedro Oliveira, and the Instituto Superior de
Educac¸
˜
ao e Ci
ˆ
encias (ISEC), for their support to this
work.
REFERENCES
Anderson, J. C., Lehnardt, J., and Slater, N. (2010).
CouchDB: The Definitive Guide Time to Relax.
O’Reilly Media, Inc., 1st edition.
Bayer, R. (1971). Binary b-trees for virtual memory. In Pro-
ceedings of the 1971 ACM SIGFIDET (now SIGMOD)
Workshop on Data Description, Access and Control,
SIGFIDET ’71, pages 219–235, New York, NY, USA.
ACM.
Brooks, D. (2009). Extensible biosignal metadata a model
for physiological time-series data. In Eng. in Medicine
and Biology Society, 2009. EMBC 2009. Annual Inter-
national Conference of the IEEE, pages 3881 –3884.
Brooks, D., Hunter, P., Smaill, B., and Titchener, M. (2011).
BiosignalML - a meta-model for biosignals. In Eng.
StorageBIT-AMetadata-aware,Extensible,SemanticandHierarchicalDatabaseforBiosignals
73

in Medicine and Biology Society,EMBC, 2011 Annual
International Conference of the IEEE, pages 5670 –
5673.
Chodorow, K. and Dirolf, M. (2010). MongoDB: The
Definitive Guide. O’Reilly Media.
Crockford, D. (2006). The application/json media type for
JavaScript Object Notation (JSON). Network Work-
ing Group, json.org/.
Dean, J. and Ghemawat, S. (2004). MapReduce: simplified
data processing on large clusters. In Proceedings of
the 6th conference on Symposium on Opearting Sys-
tems Design & Implementation - Volume 6, OSDI’04,
pages 10–10, Berkeley, CA, USA. USENIX Associa-
tion.
Goldberger, A. L., Amaral, L. A. N., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000). Phys-
ioBank, PhysioToolkit, and PhysioNet: Components
of a new research resource for complex physiologic
signals. Circulation, 101(23):e215–e220.
HDF (2010). The HDF group. Hierarchical data format ver-
sion 5, 2000-2010. www.hdfgroup.org/HDF5.
Hellmann, G., Kuhn, M., Prosch, M., and Spreng, M.
(1996). Extensible biosignal (EBS) file format: simple
method for eeg data exchange. Electroencephalogra-
phy and Clinical Neurophysiology, 99(5):426 – 431.
Jain, A., Duin, R., and Mao, J. (2000). Statistical pattern
recognition: A review. Pattern Analysis and Machine
Intelligence, IEEE Transactions on, 22(1):4–37.
Kemp, B. and Olivan, J. (2003). European data format
‘plus’ (EDF+), an edf alike standard format for the
exchange of physiological data. Clinical Neurophysi-
ology, 114(9):1755 – 1761.
Kokkinaki, A., Chouvarda, I., and Maglaveras, N.
(2008). An ontology-based approach facilitating uni-
fied querying of biosignals and patient records. In
Eng. in Medicine and Biology Society, 2008. EMBS
2008. 30th Annual International Conference of the
IEEE, pages 2861 –2864.
Kvedar, J. (2011). A physician’s perspective on self-
tracking. MIT - Technology Review.
Lovell, N., Magrabi, F., Celler, B., Huynh, K., and Gars-
den, H. (2001). Web-based acquisition, storage, and
retrieval of biomedical signals. Eng. in Medicine and
Biology Magazine, IEEE, 20(3):38 –44.
McGuinness, D. L. (2003). Ontologies come of age. Spin-
ning the Semantic Web: Bringing the World Wide Web
to Its Full Potential.
MFER (2003). Medical waveform format encoding rules.
http://ecg.heart.or.jp/En/Index.htm.
Penzel, T., Kemp, B., Klosch, G., Schlogl, A., Hasan, J.,
Varri, A., and Korhonen, I. (2001). Acquisition of
biomedical signals databases. Eng. in Medicine and
Biology Magazine, IEEE, 20(3):25 –32.
Pierce, B. C. and Vouillon, J. (2004). What’s in Unison? A
formal specification and reference implementation of
a file synchronizer. Technical Report MS-CIS-03-36,
Dept. of Computer and Information Science, Univer-
sity of Pennsylvania.
Strauch, C. (2011). NoSQL databases. Technical report,
Stuttgart Media University.
Varri, A., Kemp, B., Penzel, T., and Schlogl, A. (2001).
Standards for biomedical signal databases. Eng. in
Medicine and Biology Magazine, IEEE, 20(3):33 –37.
Vaseghi, S. V. (2006). Advanced Digital Signal Processing
and Noise Reduction. Wiley, 3rd edition.
Wal, T. V. (2007). Folksonomy coinage and definition.
http://vanderwal.net/folksonomy.html.
HEALTHINF2013-InternationalConferenceonHealthInformatics
74
