
MIST: A Tool for Rapid in silico Generation of Molecular Data from
Bacterial Genome Sequences
Peter Kruczkiewicz
1,2
, Steven Mutschall
1
, Dillon Barker
1,5
, James Thomas
2
, Gary Van Domselaar
4
,
Victor P. J. Gannon
1
, Catherine D. Carrillo
3
and Eduardo N. Taboada
1,2
1
Laboratory for Foodborne Zoonoses, Public Health Agency of Canada, Lethbridge, AB, Canada
2
Department of Biological Sciences, University of Lethbridge, Lethbridge, AB, Canada
3
Bureau of Microbial Hazards, Food Directorate, Health Canada, Ottawa, ON, Canada
4
National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada
5
Department of Microbiology, University of Manitoba, Winnipeg, MB, Canada
Keywords:
Molecular Typing, MLST, CGF, PCR, MLVA, VNTR.
Abstract:
Whole-genome sequence (WGS) data can, in principle, resolve bacterial isolates that differ by a single base
pair, thus providing the highest level of discriminatory power for epidemiologic subtyping. Nonetheless, be-
cause the capability to perform whole-genome sequencing in the context of epidemiological investigations
involving priority pathogens has only recently become practical, fewer isolates have WGS data available rela-
tive to traditional subtyping methods. It will be important to link these WGS data to data in traditional typing
databases such as PulseNet and PubMLST in order to place them into proper historical and epidemiological
context, thus enhancing investigative capabilities in response to public health events. We present MIST (Mi-
crobial In Silico Typer), a bioinformatics tool for rapidly generating in silico typing data (e.g. MLST, MLVA)
from draft bacterial genome assemblies. MIST is highly customizable, allowing the analysis of existing typing
methods along with novel typing schemes. Rapid in silico typing provides a link between historical typing
data and WGS data, while also providing a framework for the assessment of molecular typing methods based
on WGS analysis.
1 INTRODUCTION
A key component of the public health response to pri-
ority pathogens involves efforts to implement broad-
based systems for epidemiologic surveillance. These
activities are aimed at examining geographical/host
distribution and sources of disease, with the ultimate
goal of reducing the burden of illness through targeted
interventions that disrupt pathogen transmission path-
ways.
The last two decades have witnessed the emer-
gence of DNA-based subtyping methods, which have
become integral to epidemiology, making it one the
most powerful tools in public health (van Belkum,
2003). Genotyping methods have revolutionized the
field of epidemiology by enhancing the ability to dis-
criminate between isolates, which has made it invalu-
able in a range of contexts: early outbreak detection;
microbial source attribution to determine sources of
contamination and exposure; identification of the
types and subtypes of pathogenic bacteria circulating
among humans, animals, and the environment (Fox-
man and Riley, 2001). Various subtyping schemes
have been developed based on the analysis of differ-
ent types of polymorphisms that can be broadly clas-
sified into three major groups: DNA banding-pattern
based methods, such as pulsed-field gel electrophore-
sis (PFGE) and restriction fragment length polymor-
phism (RFLP); DNA hybridization-based methods,
such as PCR and microarrays; and DNA sequence-
based methods, such as multilocus sequence typing
(MLST) (Li et al., 2009). Although these methods
have been applied with varying levels of success, the
increasing resolution of modern methods has greatly
enhanced epidemiologic investigations. For example,
by identifying cases with matching subtyping data, it
has been possible to distinguish outbreak cases from
316
Krukczkiewicz P., Mutschall S., Barker D., Thomas J., Van Domselaar G., P. J. Gannon V., D. Carrillo C. and N. Taboada E..
MIST: A Tool for Rapid in silico Generation of Molecular Data from Bacterial Genome Sequences.
DOI: 10.5220/0004249003160323
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2013), pages 316-323
ISBN: 978-989-8565-35-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

concurrent sporadic cases (Singh et al., 2006).
While a few leading techniques remain widely
used tools for strain characterization, high-throughput
whole-genome sequencing (WGS) has rapidly be-
come a viable option for the investigation of prior-
ity pathogens in the context of public health events
(Gilmour et al., 2010; Alexander et al., 2012). In
principle, WGS data can resolve bacterial isolates that
differ by a single base pair, providing the highest level
of discrimination for bacterial strain characterization.
However, because the capability to perform WGS for
subtyping has only recently become practical, rela-
tively few isolates have WGS data available compared
to traditional typing methods such as PFGE or MLST,
which have dedicated databases with extensive histor-
ical data going back a decade or more. Thus, a crit-
ical component in the transition from molecular epi-
demiology based on existing genotyping approaches
to a genomic epidemiology paradigm based on the
analysis of WGS data in an epidemiologic framework
will be the linkage of WGS data to databases such as
PulseNet (Swaminathan et al., 2001) and PubMLST
(Jolley et al., 2004). This will allow genomic data to
be placed into proper historical and epidemiological
context, thus enhancing the analysis of surveillance
data and the investigation of outbreak data.
This study presents Microbial In Silico Typer
(MIST), a tool for the rapid in silico generation of
molecular typing profiles (e.g. MLST, MLVA) from
draft bacterial genome sequences. We have recently
reported results obtained using MIST as part of a
framework for assessing the performance of existing
molecular typing methods for Campylobacter jejuni
and C. coli using WGS data (Carrillo et al., 2012).
In this study we present the full implementation of
MIST and demonstrate the utility of MIST analysis
using finished, closed and draft assembly genome se-
quence data from priority human pathogens, and show
through comparison with published and experimen-
tal data that MIST produces accurate in silico-derived
molecular subtyping data for a variety of existing and
novel typing approaches.
2 IMPLEMENTATION
2.1 Overview
MIST is a standalone application written in the C#
programming language using the Microsoft .NET
Framework 4.0. MIST uses ObjectListView 2.5.0,
an open-source extension to the .NET ListView,
and DotNetZip 1.9.1.8, an open-source library for
creating and extracting ZIP files. The GUI version
WGS assembly
Microbial In Silico Typer
Probe hy-
bridization
PCR
Sequence
typing
SNP
typing
VNTR
Sequence
type
Binary
fingerprint
Hybrid-
ization
profile
SNP des-
ignation
VNTR
profile
In silico typing results
Figure 1: Overall MIST in silico typing workflow.
of MIST is currently only supported under Windows
(XP, Vista, 7). However, a command-line version
of MIST is available for both Windows and Linux.
MIST is a freely available application licensed under
the GNU General Public License. The application,
source code and documentation is available at
https://bitbucket.org/peterk87/microbialinsilicotyper
and https://bitbucket.org/peterk87/mist for the GUI
and command-line versions of MIST, respectively.
All analyses performed within MIST utilize se-
quence similarity searching, which is performed
using blastn from the NCBI BLAST+ application
suite (Altschul et al., 1990; Camacho et al., 2009).
MIST can produce in silico data for PCR-, probe
hybridization-, sequence-, variable number of tandem
repeat- (VNTR), and SNP-based typing assays (Fig-
ure 1) while also providing users with detailed sum-
mary data underlying the subtyping results.
MIST was designed such that the user is free to
use existing typing schemes or to create their own.
Novel schemes can be based on conventional sub-
typing approaches or can incorporate molecular tar-
gets that would be impractical in the context of a
laboratory-based assay. For a given bacterial species,
an existing Package, consisting of one or more typing
schemes or Assays, themselves comprised of one or
more Markers, can be loaded and executed in MIST,
yielding in silico results for the assays selected. An
assay creation module allows the user to easily edit or
create new packages and assays through an entry form
or through the import of a tab-delimited file with the
necessary assay parameters.
MIST allows the user to easily export typing re-
sults or to save and continue an analysis with addi-
tional WGS data or typing schemes.
MIST:AToolforRapidinsilicoGenerationofMolecularDatafromBacterialGenomeSequences
317

2.2 PCR-based Typing
WGS assembly
BLAST search
PCR primers
Primer binding sites
Find most likely amplicon
Correct
amplicon
size?
Present Absent
Determine copy number
Yes No
VNTR-based typing
Figure 2: In silico VNTR- and PCR-based typing workflow.
MIST simulates in silico PCR using BLAST se-
quence similarity searching (Figure 2). BLAST
searches are conducted to find all potential for-
ward and reverse primer binding sites in the subject
genome. All combinations of potential forward and
reverse primer sites are investigated to determine pu-
tative amplicons. A positive result is reported when
an amplicon can be retrieved within the expected size
tolerances and in which the amplicon is flanked by
the forward and reverse primers in either correct ori-
entation. If all primer binding site combinations have
been exhausted and no suitable amplicon is gener-
ated, a negative result is reported. Although tradi-
tional in vitro PCR assays produce only a binary sig-
nal, MIST provides a more in-depth analysis of PCR
results. A summary of binary absence-presence re-
sults along with detailed match information can be
accessed for each PCR marker match.
2.3 VNTR-based Typing
MIST simulates in silico VNTR results (i.e. MLVA)
by using a variation on in silico PCR analysis (Fig-
ure 2). If an amplicon can be retrieved with a length
less than the user-defined maximum amplicon size
threshold (3000 bp by default), the copy number or
the number of repeats is reported. If a suitable am-
plicon cannot be found, a null result is returned. Al-
though the copy number reported is typically zero or
an integer, in the case of partial repeats a non-integer
number may be reported. To aid in the intrepretation
of the data, detailed match information, such as the
most likely primer binding sites and predicted ampli-
con, is made available.
2.4 Probe Hybridization-based Typing
WGS assembly
BLAST search
Probe
BLAST hit
above the
IDT?
AbsentPresent
Determine SNP designation
Yes No
SNP-based typing
Figure 3: In silico probe hybridization- and SNP-based typ-
ing workflow.
Two modes for in silico probe hybridization anal-
ysis are available in MIST. ‘Mode 1’, which is the
default setting, is for ‘short’ probes (i.e. < 70 bp or
user-defined), such as those which might be found on
oligonucleotide-based microarrays. ‘Mode 2’ is for
‘long’ probes (i.e. > 70 bp or user-defined), such as
those which might be found in amplicon-based mi-
croarrays. For ‘short’ probes the hybridization profile
for a genome is predicted using BLAST based on a
user-defined percent identity threshold (IDT) for each
probe in the assay. For ‘long’ probes, an alignment
length threshold (ALT) is used in addition to the IDT.
In both modes, BLAST is used to identify hits for
each probe; the hit with highest bit score is used if
multiple BLAST hits can be identified. In Mode 1,
a positive hit is reported if the top BLAST hit has a
percent identity equal to or greater than the IDT over
the entire length of the probe. If no BLAST hits can
be identified or if the top hit does not fulfill the IDT
criterion, a negative result is reported. In Mode 2, a
positive hit is reported if the top BLAST hit has a per-
cent identity and alignment length equal to or greater
than the IDT and ALT, respectively. If no BLAST hits
can be identified or if the top hit does not fulfill the
IDT and ALT criteria, a negative result is reported. In
addition to the binary absence-presence results MIST
provides detailed match information, which is also
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
318
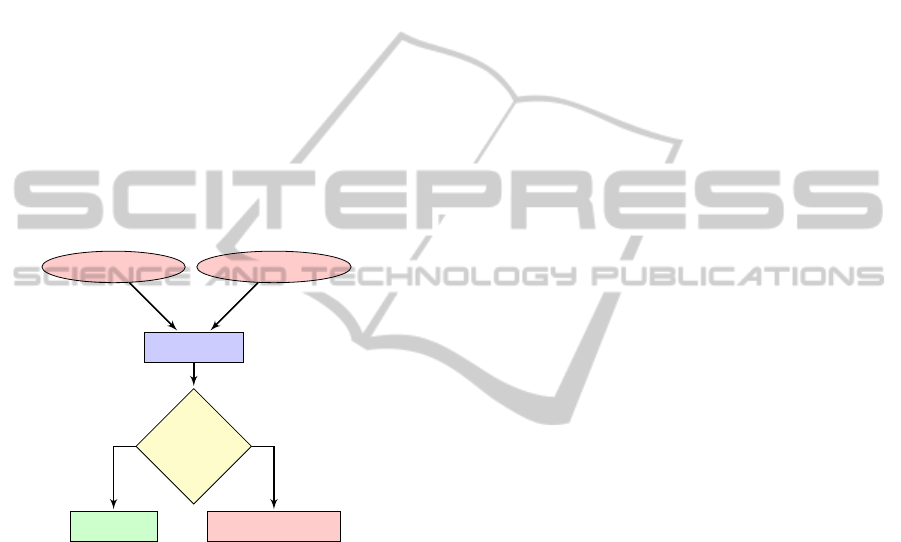
provided for negative results, thus allowing the user
to verify results obtained.
2.5 SNP-based Typing
SNP loci can be interrogated using a variation of
the in silico probe hybridization (Figure 3). To de-
sign a SNP-based assay, the SNP position is indi-
cated by an ‘N’ within a probe sequence. Probe se-
quences are queried against the subject genome using
BLAST. The nucleotide at a SNP position is reported
if a match above the IDT can be found in the sub-
ject genome. Other match information made avail-
able includes the probe match sequence and the loca-
tion within the subject genome. If a match above the
IDT cannot be found within the subject genome, the
top BLAST hit is provided along with the number of
mismatches with respect to the query probe sequence.
2.6 Sequence-based Typing
WGS assembly
BLAST search
Allele sequences
Perfect
match
found?
Allele match Closest allele match
Yes No
Figure 4: In silico sequence-based typing workflow.
The allelic profile of a genome for a given gene
marker is determined through sequence similarity
searching of a database of the known alleles for the
gene using BLAST (Figure 4). MIST utilizes a rapid
search strategy wherein one allele of each unique size
for each allele database is queried against the subject
genome. A simple matching algorithm is then used to
find a perfect match in the allelic database. If a perfect
match cannot be found using this approach, complete
pairwise BLAST searching against the allele database
is performed to determine the closest matching alle-
les (i.e. the alleles with least number of mismatches)
along with the number of SNPs with respect to the
subject genome’s allele.
In addition to the individual allelic profiles, MIST
automatically determines the overall sequence type
(ST) and clonal complex (CC). If additional metadata
is linked to allelic profiles, this ancillary data can be
displayed along with ST and CC information.
3 RESULTS AND DISCUSSION
Wherever possible, MIST-derived typing results were
validated by comparison against experimentally-
derived typing results obtained from the literature.
The accuracy of MIST is inherently linked to the
quality of the WGS sequence data (i.e. the subject
genome) used as an input because genome assemblies
with large numbers of contigs, or regions of poor or
absent sequence data may introduce false negatives
to the in silico results (Carrillo et al., 2012). Simi-
larly, accuracy can be impacted by the error rate in
experimental data, which is largely due the inherent
noise in hybridization, a process upon which most
subtyping methods rely. Nevertheless, MIST was
found to generate sensitive analyses and in silico re-
sults that were highly concordant with experimental
data whether from completed whole genome data or
from minimally processed draft genome assemblies.
3.1 PCR-based Typing: Campylobacter
jejuni CGF40 Assay
The comparative genomic fingerprinting (CGF) assay,
CGF40 (Taboada et al., 2012), was used to assess the
performance of in silico PCR analysis using MIST.
The CGF40 assay targets 40 accessory genes present
within C. jejuni NCTC 11168 and variably present in
other C. jejuni and C. coli strains. The 40 primer pairs
along with the expected amplicon sizes and an ampli-
con size tolerance of 5% were input into MIST as an
Assay.
Based on MIST CGF40 analysis of 25 sequenced
C. jejuni genomes, the average concordance between
in silico and in vitro-derived results was found to be
92.8% (928/1000 matches) using a blastn word size
of 8. The false negative and false positive rates were
4.0% and 3.2%, respectively. Generally, false nega-
tive results were attributable to sequence truncations
within incomplete draft assemblies. False positive
results were largely “true” amplicons that contained
SNPs within priming sites, which would likely affect
amplification under standard PCR conditions. For ex-
ample, marker cj1721 yielded a concordance of only
48%, with 13 false positive discrepancies. A multi-
ple sequence alignment of the retrieved cj1721 am-
plicons revealed several SNPs in both priming sites.
However, after reducing the stringency of the test by
increasing the blastn word size, the false positive rate
for cj1721 dropped from 48% to 12% without ap-
preciably changing the overall concordance of the re-
maining markers in the assay.
Thus, the blastn word size can be used to adjust
the stringency of an in silico PCR test. A larger word
MIST:AToolforRapidinsilicoGenerationofMolecularDatafromBacterialGenomeSequences
319

size (e.g. >7) will produce results that are highly
specific (and less sensitive) with respect to the given
primer inputs; likely producing results that are more
concordant with in vitro PCR due to fewer false pos-
itive matches. Alternatively, a lower word size (e.g.
<7) will produce results that are more sensitive (and
less specific), which may be useful for exploratory
analyses where multiple sequence alignments of the
retrieved amplicons can be investigated to further ex-
amine differences between strains.
MIST offers user control over the in silico results
and provides a variety of output data making it a valu-
able tool in the validation and creation of novel PCR
typing schemes.
3.2 VNTR-based Typing: Listeria
monocytogenes MLVA
VNTR typing was simulated using an MLVA scheme
for L. monocytogenes (Miya et al., 2008) on 23 pub-
licly available L. monocytogenes genomes. In silico
VNTR data generated by MIST are shown in Table 1.
The in silico VNTR results for F2365 matched the ex-
pected results with 14, 17 and 9 repeats in the VNTR
loci TR1, TR2 and TR3, respectively.
It was not possible to validate results obtained
by MIST in any L. monocytogenes strains other than
F2365 because of the lack of available MLVA data on
genome-sequenced strains in the literature for this or
other MLVA schemes. MIST was unable to find an
amplicon in some cases for the MLVA marker TR1.
This is consistent with previous observations on the
potential unsuitability of the TR1 locus due to vari-
ability between strains in that region (Miya et al.,
2008). Our results thus illustrate how MIST may be
a valuable tool in the in silico validation and develop-
ment of novel VNTR-based typing schemes.
3.3 Probe Hybridization-based Typing:
Escherichia coli Pathotyping
Microarray
Probe hybridization was simulated using the probe se-
quences from the E. coli miniaturized pathotyping mi-
croarray (Anjum et al., 2007) on 110 E. coli genomes
obtained from NCBI. The microarray consists of a
combination of 61 probes targeting virulence, bacte-
riocin and conserved genes that are shared among or
specific to E. coli pathotypes. Concordance was cal-
culated between the in silico and in vitro array for the
seven E. coli strains shared between the public and
study datasets.
The overall concordance for these seven E. coli
Table 1: The number of predicted repeats for the in silico
L. monocytogenes MLVA scheme (Miya et al., 2008) are
shown along with the lineage of each strain. In silico MLVA
data for F2365 is in boldface. In some cases with TR1,
MIST was unable to find a suitable amplicon (missing data
indicated with (–)).
Strain Lineage TR1 TR2 TR3
F2365 I 14 17 9
J1-194 I 6 18 5
N1-017 I 15 46.83 6
R2-503 I 13 46.83 6
Clip80459 I 15 19 5
H7858 I 23 22 5
HPB2262 I – 12 7
Scott-A I 14 17 9
F6854 II 18.22 8.33 9
F6900 II 21 9.5 9
J0161 II 15.56 7.83 9
J2818 II 21 9.5 9
N3-165 II 16.11 8.33 9
1988 II – 8.33 9
10403S II 7.33 8 9
EGD-e II 26.44 11.5 9
R2-561 II 17.67 8 9
08-5923 II 35 12 9
08-5578 II 35 12 9
HCC23 III – 22 5
L99-4a III – 22 5
M7 III – 22 5
J2-071 III – 16 5
strains was 93.4% (399/427 matches) using a probe
percent identity threshold of 90%. The false nega-
tive and false positive rates were 2.8% and 3.8%, re-
spectively. The rrl 23S rRNA control gene probes
accounted for the majority of discrepancies (71.4%),
since four probes from this locus had mismatches in
more than half of the strains tested. Removal of the
rrl control gene probes increased the concordance to
97.9% (322/329 matches). Generally, false negative
results by MIST were attributable to SNPs within the
probe hybridization site resulting in a %ID below the
90% ID threshold. False positive results by MIST
were confirmed to be full length or near full length
matches with greater than 95%ID (0-2 mismatches).
In silico pathotype designations are shown in Table 2.
Table 2: The in silico E. coli pathotyping microarray
analysis-derived pathotypes for 7 E. coli strains.
Sample Pathotype
536 ExPEC
IHE3034 ExPEC
K-12 MG1655 –
B171 EPEC;EHEC
CFT073 EAEC;ETEC;ExPEC
O127:H6 E2348-69 EAEC;ETEC;EPEC;EHEC
O157:H7 EDL933 EAEC;ETEC;EPEC;EHEC;STEC
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
320

3.4 SNP-based Typing: Listeria
monocytogenes TMLGT
An in silico SNP-based typing assay was created
based on the targeted multilocus genotyping assay
(TMLGT) for L. monocytogenes (Ward et al., 2010).
The 30 probe assay simultaneously classifies the 4
lineages, 4 major serotypes and four epidemic clones
(EC) of L. monocytogenes by interrogating a 3’ SNP
diagnostic for one of the aforementioned groups. The
MIST assay for TMLGT was tested against 23 pub-
licly available L. monocytogenes genome sequences
and validated by comparing known lineage, serotype
and EC designations to the MIST-derived designa-
tions. MIST results for the TMLGT assay placed all
23 strains into their correct lineages. Serotypes were
correctly assigned to 15 out of 17 strains belonging to
one of the four serotypes targeted by this assay. All
eight strains belonging to known EC groups were cor-
rectly identified (Table 3).
SNP-based typing assays are particularly
amenable to analysis in MIST because the results
require little analytical interpretation, as a SNP vari-
ant determination is directly linked to the underlying
sequence data. Furthermore, because the assay is
performed in silico, literally any number of variant
sites can be tested for, allowing for the development
and testing of assays consisting of hundreds if not
thousands of markers, which would be difficult to
deploy in a laboratory setting.
3.5 Sequence-based Typing:
Staphylococcus aureus MLST
Sequence-based typing was simulated using the
MLST scheme for Staphylococcus aureus (Enright
et al., 2000). Genome sequences for 63 publicly
available S. aureus strains for which correspond-
ing STs were available from the MLST database
(saureus.mlst.net) were examined. The concordance,
calculated by computing the number of matching al-
leles between in vitro and in silico MLST data as a
proportion of the total number of alleles, was found
to be 98.6% (435/441 matching alleles).
Six allelic discrepancies were found among five
genomes, and were investigated using the detailed
match information available within MIST. Five dis-
crepancies were cases in which a different allele num-
ber was assigned based on a minor SNP variant; four
cases differed by a single SNP, and one case differed
by two SNPs. In each case, results obtained with
MIST reflected a canonical match between the un-
derlying WGS data provided and the discordant al-
lele in the database. For the remaining case of allelic
Table 3: The in silico TMLGT analysis-derived lineage,
serotype and EC designations of 23 L. monocytogenes
strains are shown here. In silico-derived data not matching
in vitro-derived data are shown in brackets.
Sample Lineage Serotype EC
F2365 I 4b I
J1-194 I 1/2b –
N1-017 I (1/2b) –
R2-503 I 1/2b –
Clip80459 I 4b –
H7858 I 4b II
HPB2262 I 4b Ia
Scott-A I 4b Ia
F6854 II 1/2a III
F6900 II 1/2a III
J0161 II 1/2a III
J2818 II 1/2a III
N3-165 II 1/2a –
1988 II 1/2a –
10403S II 1/2a –
EGD-e II (1/2c) –
R2-561 II 1/2c –
08-5923 II 1/2a –
08-5578 II 1/2a –
HCC23 III – –
L99-4a III – –
M7 III – –
J2-071 III – –
discrepancy, the allele for arcC in genome TCH60
could not be identified. Further examination revealed
a single-base adenine deletion within a 7-base poly-
adenine tract; an error common in 454 pyrosequenc-
ing. After correcting for this assumed error, the
TCH60 arcC allele was correctly assigned by MIST
as arcC 2. These data not only show that MIST can
accurately determine MLST data with a very high de-
gree of confidence, but also illustrate how the detailed
information provided by MIST can prove useful.
The MIST platform could be used in the testing
and development of expanded MLST schemes includ-
ing whole-genome MLST analysis, which are of great
interest in the context of rapid whole genome-based
subtyping of strains. Moreover, the provision of ad-
ditional subtype-associated metadata is a feature not
common among other in silico MLST tools, includ-
ing those provided by online MLST databases.
4 CONCLUSIONS
The current generation of molecular typing meth-
ods was originally conceived as a proxy for whole-
genome sequence data at a time when whole genome
sequencing had not yet been achieved, or more re-
cently, when the cost and logistics precluded the se-
MIST:AToolforRapidinsilicoGenerationofMolecularDatafromBacterialGenomeSequences
321

quencing of more than one or a small number of
strains for a particular species.
Ultimately, high quality, whole genome sequenc-
ing is the true gold standard for molecular character-
ization of microbes. In addition to encoding all of
the information necessary to determine conventional
molecular typing profiles, WGS provides the most
comprehensive data for inferring strain to strain phy-
logenetic relationships. For these reasons, WGS anal-
ysis has become increasingly prominent in the public
health response to pathogens, enabling characteriza-
tion of organisms at an unprecedented level of reso-
lution. Moreover, as the cost of sequencing continues
to decline, the applicability of WGS will broaden to
a larger scope of strains and, in the near future, will
likely become the method of choice for characteriza-
tion of all microbes.
In the context of public health, a significant gap
currently exists in that although WGS analysis has
been shown to be viable in the context of public health
events such as outbreaks (Gilmour et al., 2010), it may
not yet be possible to perform WGS in the context of
routine epidemiologic surveillance. In silico typing
provides the opportunity for the utilization of WGS
data as a framework for comparing the performance
of existing molecular typing methods or for assessing
and validating novel methods (Carrillo et al., 2012).
Moreover, while ever-expanding, WGS datasets
still comprise only a fraction of the historical data
contained in public repositories of molecular typing
data, such as PulseNet (Swaminathan et al., 2001) and
PubMLST (Jolley et al., 2004), and this is not likely
to change in the immediate future. Establishing links
between WGS data and the data contained in molecu-
lar epidemiology databases will be critical as we tran-
sition from a paradigm involving molecular typing to
one that is based on WGS.
Recently developed comparative genomics-based
tools have been developed with the aforementioned
goals in mind. A web-based tool was recently devel-
oped for in silico MLST analysis of bacterial isolates
using WGS data (Larsen et al., 2012). Bacterial Iso-
late Genomic Sequence Database (BIGSDB), which
is based on the mlstdbNet platform (Jolley et al.,
2004), has been developed for storage and analysis
of WGS data along with other types of data such as
MLST data (Jolley and Maiden, 2010). BIGSDB has
been used in conjunction with Ion Torrent sequenc-
ing to rapidly generate accurate typing data on multi-
ple methods for Neisseria meningitidis (Vogel et al.,
2012). MIST was designed to provide a complimen-
tary approach to existing tools by allowing users the
flexibility to analyze draft WGS data and produce in
silico typing profiles for traditional typing schemes,
assays based on modifications on these traditional
schemes, and assays based on novel typing schemes.
In silico typing profiles can be linked to histor-
ical typing data from public databases and to other
available metadata on each genome analysed, such
as phenotypic, clinical, or epidemiological informa-
tion, enabling the user to examine associations be-
tween the sub-typing data and their underlying meta-
data. Since MIST allows the user to design and test
schemes based on approaches that may be imprac-
tical to perform in vitro, extended MLST schemes
or hybridization-based methods targeting hundreds of
loci can be tested that would otherwise be subject
to sensitivity/specificity issues when deployed in the
lab. Furthermore, the MIST platform enables compar-
ison between typing methods through the simultane-
ous generation of in silico typing results for a variety
of different methods.
As we move into the era of broad-based WGS,
it is important to consider the relationship between
WGS data and historical molecular typing databases,
which are rich sources of molecular and epidemiolog-
ical information. Ideally, these sources of information
should be used cooperatively; large molecular epi-
demiological databases should be used as resources
for targeted selection of strains for WGS projects,
and the corollary, as WGS begins to replace to tra-
ditional genotyping, the ability to rapidly assess rel-
evant genetic markers from an abundance of mini-
mally processed sequence data will become an es-
sential requirement towards leveraging these data for
maximum public health impact. Continued efforts to-
ward the development of bioinformatics tools that en-
able rapid analysis of draft genome data will enable
public health laboratories to not only link WGS data
to historical data but provide a framework for use in
the development of novel analytical methods and pro-
vide an improved understanding of the performance
of current methods to assist in the interpretation of
existing studies.
REFERENCES
Alexander, D. C., Hao, W., Gilmour, M. W., Zittermann,
S., Sarabia, A., Melano, R. G., Peralta, A., Lombos,
M., Warren, K., Amatnieks, Y., Virey, E., Ma, J. H.,
Jamieson, F. B., Low, D. E., and Allen, V. G. (2012).
Escherichia coli O104:H4 infections and international
travel. Emerging Infectious Diseases, 18(3):473–476.
PMID: 22377016.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and
Lipman, D. J. (1990). Basic local alignment search
tool. Journal of Molecular Biology, 215(3):403–410.
Anjum, M. F., Mafura, M., Slickers, P., Ballmer, K., Kuh-
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
322

nert, P., Woodward, M. J., and Ehricht, R. (2007).
Pathotyping Escherichia coli by using miniaturized
DNA microarrays. Applied and Environmental Mi-
crobiology, 73(17):5692–5697.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Pa-
padopoulos, J., Bealer, K., and Madden, T. L. (2009).
BLAST+: architecture and applications. BMC Bioin-
formatics, 10:421. PMID: 20003500.
Carrillo, C. D., Kruczkiewicz, P., Mutschall, S., Tudor, A.,
Clark, C., and Taboada, E. N. (2012). A framework for
assessing the concordance of molecular typing meth-
ods and the true strain phylogeny of Campylobac-
ter jejuni and C. coli using draft genome sequence
data. Frontiers in Cellular and Infection Microbiol-
ogy, 2:57.
Enright, M. C., Day, N. P. J., Davies, C. E., Peacock, S. J.,
and Spratt, B. G. (2000). Multilocus sequence typ-
ing for characterization of methicillin-resistant and
methicillin-susceptible clones of Staphylococcus au-
reus. Journal of Clinical Microbiology, 38(3):1008–
1015.
Foxman, B. and Riley, L. (2001). Molecular epidemiology:
focus on infection. American Journal of Epidemiol-
ogy, 153(12):1135–1141. PMID: 11415945.
Gilmour, M. W., Graham, M., Domselaar, G. V., Tyler,
S., Kent, H., Trout-Yakel, K. M., Larios, O., Allen,
V., Lee, B., and Nadon, C. (2010). High-throughput
genome sequencing of two Listeria monocytogenes
clinical isolates during a large foodborne outbreak.
BMC Genomics, 11(1):120.
Jolley, K. A., Chan, M., and Maiden, M. C. (2004). mlstdb-
Net - distributed multi-locus sequence typing (MLST)
databases. BMC Bioinformatics, 5:86–86. PMID:
15230973 PMCID: 459212.
Jolley, K. A. and Maiden, M. C. J. (2010). BIGSdb: scal-
able analysis of bacterial genome variation at the pop-
ulation level. BMC Bioinformatics, 11:595. PMID:
21143983.
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Has-
man, H., Marvig, R. L., Jelsbak, L., Sicheritz-Pont
´
en,
T., Ussery, D. W., Aarestrup, F. M., and Lund, O.
(2012). Multilocus sequence typing of total-genome-
sequenced bacteria. Journal of Clinical Microbiology,
50(4):1355–1361. PMID: 22238442.
Li, W., Raoult, D., and Fournier, P. (2009). Bacterial strain
typing in the genomic era. FEMS Microbiology Re-
views, 33(5):892–916. PMID: 19453749.
Miya, S., Kimura, B., Sato, M., Takahashi, H., Ishikawa, T.,
Suda, T., Takakura, C., Fujii, T., and Wiedmann, M.
(2008). Development of a multilocus variable-number
of tandem repeat typing method for Listeria mono-
cytogenes serotype 4b strains. International Jour-
nal of Food Microbiology, 124(3):239–249. PMID:
18457891.
Singh, A., Goering, R. V., Simjee, S., Foley, S. L., and
Zervos, M. J. (2006). Application of molecular tech-
niques to the study of hospital infection. Clinical Mi-
crobiology Reviews, 19(3):512–530.
Swaminathan, B., Barrett, T. J., Hunter, S. B., and Tauxe,
R. V. (2001). PulseNet: the molecular subtyp-
ing network for foodborne bacterial disease surveil-
lance, united states. Emerging Infectious Diseases,
7(3):382–389. PMID: 11384513.
Taboada, E. N., Ross, S. L., Mutschall, S. K., Mackinnon,
J. M., Roberts, M. J., Buchanan, C. J., Kruczkiewicz,
P., Jokinen, C. C., Thomas, J. E., Nash, J. H. E.,
Gannon, V. P. J., Marshall, B., Pollari, F., and Clark,
C. G. (2012). Development and validation of a
comparative genomic fingerprinting method for high-
resolution genotyping of Campylobacter jejuni. Jour-
nal of Clinical Microbiology, 50(3):788–797. PMID:
22170908.
van Belkum, A. (2003). High-throughput epidemiologic
typing in clinical microbiology. Clinical Microbiol-
ogy and Infection, 9(2):86–100.
Vogel, U., Szczepanowski, R., Claus, H., J
¨
unemann, S.,
Prior, K., and Harmsen, D. (2012). Ion torrent per-
sonal genome machine sequencing for genomic typ-
ing of Neisseria Meningitidis for rapid determination
of multiple layers of typing information. Journal of
Clinical Microbiology, 50(6):1889–1894.
Ward, T. J., Usgaard, T., and Evans, P. (2010). A targeted
multilocus genotyping assay for lineage, serogroup,
and epidemic clone typing of Listeria monocytogenes.
Appl. Environ. Microbiol., 76(19):6680–6684.
MIST:AToolforRapidinsilicoGenerationofMolecularDatafromBacterialGenomeSequences
323
