
Evaluation Methodology for Descriptors in Neuroimaging Studies
M. Luna
1,2
, F. Gayá
1
, C. Cáceres
1,2
, J. M. Tormos
3
and E. J. Gómez
1,2
1
Bioengineering and Telemedicine Centre, ETSI de Telecomunicación, Universidad Politécnica de Madrid, Madrid, Spain
2
Biomedical Research Networking Center in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN),
Zaragoza, Spain
3
Institut Guttmann, Neurorrehabilitation Hospital, Badalona, Spain
Keywords: Neuroimaging, Detection, Descriptor, Landmark, Evaluation Methodology.
Abstract: Automatic identification and location of brain structures is one of the main stages to process neuroimaging
studies. The proposed approach consists of identifying landmarks over an image. These landmarks must
have values of location and intensity variation to obtain a direct relation between detected landmarks and
brain structures. Descriptors are algorithms whose function is to select and store points featuring these two
types of information. There are many algorithms used to obtain descriptors. Therefore, it is necessary to
select the most adequate to the type of images and context of application. It is advisable to design and
develop an evaluation methodology to objectively identify appropriate algorithms. This paper proposes a
new evaluation methodology for descriptors used on neuroimaging studies.
1 INTRODUCTION
Identification and location of brain structures is a
main stage to process neuroimaging studies. One
approach consists of detecting points over the image
whose characteristics of location and intensity
permit to find a direct relationship between them and
an anatomic brain structure. These image points are
called landmarks.
Different research groups have developed
methods for detecting landmarks on neuroimaging
studies in the last years. There are two main types of
methods: semiautomatic (Izard et al., 2005),
(Shattuck et al., 2009), which require the interaction
of the user and automatic (Verard et al., 1997), (Lui
et al., 2006).
The automatic detection and identification of
landmarks allows increasing the current knowledge
about anatomic alterations and reducing the cost of
time spent by a specialist due to the fact that they
have to manually label these areas on a volumetric
image study.
An approximation based on descriptors to detect
landmarks is proposed on (Luna et al., 2012); they
present an analysis of the applicability of descriptors
to identify landmarks that have a relation with brain
structures on MRI studies. A descriptor is an
algorithm aiming to detect points that present
singular characteristics to be identified among its
neighbours. Main algorithms are SIFT (Scale
Invariant Feature Transform) (Lowe, 1999) and
SURF (Speeded Up Robust Feature) (Bay et al.,
2008).
In order to find the relation between landmarks
and brain structures it is necessary to detect
homologous pairs of points between the descriptors
of patient’s image study and the image study
containing information about brain structures of
interest. The definition of pair of landmarks changes
depending on the implemented approach of
matching and the type of distance between
descriptors (Euclidean’s distance and Mahalanobis
distance). These approaches will be described in
Methods section. In this application context, the
relation between points should be unique, namely,
there can be only a valid correspondence between
landmarks of the subject image and template image.
Therefore, the function used to identify and match
homologous points has to be biyective.
Landmarks used to identify brain structures have
to fulfill with these conditions: compromise between
processing time and number of pairs of homologous
points detected; sample's representativeness over the
region of interest (this region represents about 45%
of the image's area); and stability towards changes
on the image.
114
Luna M., Gayá F., Cáceres C., Tormos J. and Gómez E..
Evaluation Methodology for Descriptors in Neuroimaging Studies .
DOI: 10.5220/0004298001140117
In Proceedings of the International Conference on Computer Vision Theory and Applications (VISAPP-2013), pages 114-117
ISBN: 978-989-8565-48-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
So as to integrate descriptor algorithms into image
processing systems it is necessary to introduce new
changes on them. These changes will permit to
improve the current relation between processing
time and identified brain structures. Our research
group is currently developing new methods
including changes. In literature, there is any
evaluation methodology whose aim is to evaluate
objectively these algorithms on neuroimaging
studies. The main aim of this paper is to design an
evaluation methodology to compare descriptors for
detecting brain structures on neuroimaging studies.
2 MATERIALS AND METHODS
2.1 Materials
The materials used to evaluate descriptors
algorithms is firstly a set of images, on our
application context will be magnetic resonance
images. Main differences on MRI images are caused
by changes of vision angle and scale. Then, two sets
of scaled and rotated images with different angles
are necessary.
In order to look for anatomical structures, a
template image in which the brain structures appear
manually segmented is created. In this template
study, a RGB label, centre and area of the region of
interest are assigned to each brain structured.
2.2 Methods
Descriptors of each image involved on the
evaluation are obtained by applying different
algorithms. Afterwards, pairs of homologous points
between descriptors are found.
As mentioned before, there are four strategies to
identify a pair of homologous points. The first one
considers a pair of homologous points only if the
distance between descriptors is below a threshold. In
this case, several correspondences among points can
appear and several of them may be correct. The
second one identifies the nearest neighbour and
imposes a threshold. With this approach, there is
only one correspondence between points. Thus, the
relation is biyective. The third matching approach is
similar to the last one, but it estimates the distance
ratio between the first and the second nearest
neighbour and applies a threshold to this ratio (1).
μ
<
−
−
20
10
DD
DD
(1)
Where D0 is the point of interest, D1 is the first
nearest neighbour and D2 is the second nearest
neighbour; and σ is the threshold.
Based on these three approaches described on
(Mikolajczk et al., 2005) (threshold based matching,
nearest neighbor matching and nearest neighbor
distance ratio matching), this paper proposes a
fourth approach. This new approach takes into
account the fact that two types of different
information are necessary for considering two pair
of points as homologous: location and intensity
values. Then, a pair of landmarks will be considered
as homologous only if the normalized spatial
distance and the normalized descriptor distance are
minimal and stay below a threshold defined for each
distance. Both thresholds will be determined taking
into account the size of the images and the average
intensity changes detected. This approach obtains a
biyective matching function. Both distances are
balanced independently to evaluate descriptors with
these two parameters and obtain more restrictive
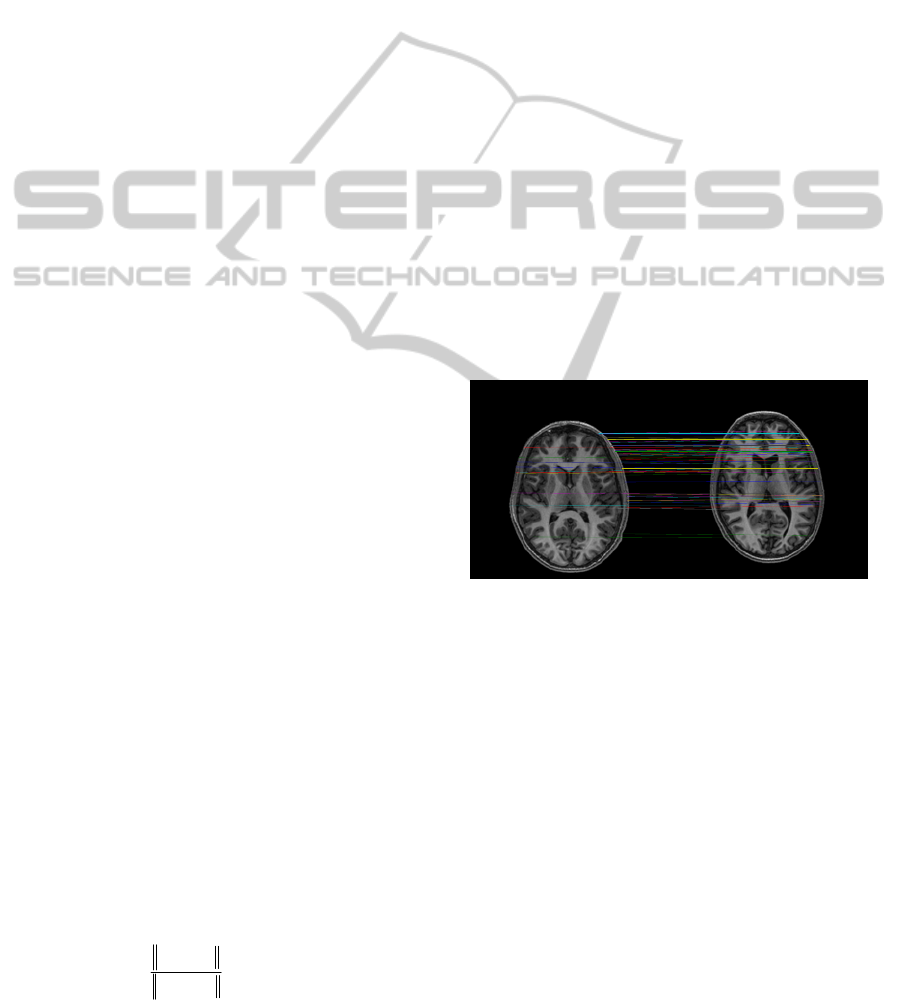
results than previous approaches. An example of
pair of homologous points detected is showed in
Figure 1. These landmarks have been obtained by
using SIFT algorithm. As can be observed, most of
detected landmarks are located over skull.
Figure 1: Pairs of homologous points.
In order to evaluate the stability of descriptors
against scaled and rotated images it is necessary to
obtain the average pairs of homologous landmarks
between original images and changed images.
Therefore, it is necessary to obtain two sets of
images, as mentioned before, scaled images and
rotated images. The fourth matching approach is
used to obtain the pairs of homologous points. An
example is showed in Figure 2. In this figure, a set
of homologous points obtained by SURF descriptor
is obtained on rotated images (top image) and
obtained by an own algorithm on scaled images
(bottom image). As can be observed, SURF
algorithm presents a similar problem as SIFT,
detected landmarks appear around skull and
longitudinal fissure. However, our algorithm obtains
landmarks also on internal structures.
EvaluationMethodologyforDescriptorsinNeuroimagingStudies
115

Homologous landmarks detected
Figure 2: Pairs of homologous points on images with
rotated changes (top) and scaled changes (low).
3 EVALUATION
METHODOLOGY
Evaluation methods will be classified into two
different sets: a general test set, whose aim is to
evaluate the descriptor's efficiency over any type of
images (in our case medical images); and a specific
test set, whose aim is to evaluate the descriptor's
efficiency to find brain structures using detected
landmarks.
The analysis parameters are: mean processing
time, average of pair of homologous points detected,
descriptor's stability considering scale changes and
rotated images and average sample’s
representativeness per area and per brain structure of
interest.
3.1 General Test Set
This set of tests evaluates processing time,
performance, pairs of homologous points detected,
stability of descriptors with image changes and
sample’s representativeness per area.
The processing time and the number of
homologous points are obtained per descriptor. A
low value of processing time and a large pairs of
homologous points are desirable.
The descriptor performance is tested by using
this parameter (Gossow et al., 2011): recall. This is
the number of correct found matches relative to the
total number of found matches (2).
matchestotal
matchestrue
recall
_
_
=
(2)
The stability of descriptors towards changes on
the image is estimated by obtaining the average of
pairs of homologous points between original and
change images and by analysing the number of true
positives detected. The higher the number of true
positives is, the bigger the descriptor’s stability.
Two possible approaches can be used to evaluate
the sample’s representativeness per area. The first
one requires obtaining the mask of the region of
interest of the image and the total area of this region.
A ratio between the number of detected landmarks
(true positives) over this region and total area of the
region is calculated (3). The closer to the unit this
parameter is the better sample’s distribution.
areatotal
landmarkstectedde
tivenessrepresenta
_
_.
=
(3)
The second approach permits to evaluate
landmarks distribution homogeneity over the region
of interest. Based on Delaunay’s triangulation using
detected landmarks, it calculates the area of these
triangles and the variance of these areas. A low
value of variance means that all of these triangles
have similar area values. Thus, the detected
landmarks present a uniformity distribution over the
area of interest.
3.2 Specific Test Set
These set of tests permit to find the relation between
detected landmarks and brain structures of interest.
The template image is used among different test.
Our approach to find the landmarks presenting a
relation with brain structures consists on seeking
through the descriptor, a point will be or not selected
as landmarks taking into account location and
descriptor information.
Sample’s representativeness per brain structure
and descriptor’s efficiency are the parameters used
to quantitatively analyse the obtained results. Brain
structures of interest can be located around cortical
and subcortical areas, so it is necessary to obtain
landmarks on both areas. The first parameter permits
to obtain the number of landmarks that can identify
each brain structure of interest, so it evaluates
whether or not the algorithm detects landmarks on
cortical or subcortical areas. Efficiency is defined as
the ratio between number of landmarks presenting a
relation with brain structures and the total number of
detected landmarks (4). The closer to the unit this
parameter is, the more useful to detect brain
structures are the detected landmarks.
landmarksntotal
landmarksbrainn
efficiency
__
__
=
(4)
VISAPP2013-InternationalConferenceonComputerVisionTheoryandApplications
116

4 RESULTS
This section describes briefly some results obtained
with SIFT and SURF algorithm to validate the
methodology proposed. A set of 10 healthy subjects,
with an age range 19-30 years, have been used to
obtain these results.
Regarding general test set, Table 1 sums up the
results obtained. Table 2 summarizes the results
obtained by specific test set. Five brain structures
have been selected to obtain these evaluation
parameters
Table 1: Summary of general test set.
SIFT SURF
Processing
time
1,45 (1,16-1,71) 2,02 (1,89-2,08)
Homologous
landmarks
732 (685-773) 1154 (951-1380)
Performance 43% 47%
Stability
2º 5º 10º 2º 5º 10º
63% 53% 43% 56% 52% 49%
Table 2: Summary of specific test set.
Sample's representativeness
SIFT SURF
Cave of Septum Pellucidum
2(1-2) 2(1-2)
Superior Sagital Sinus
7(1-6) 8(4-10)
Chroid Plexus
3(2-4) 3(3-4)
Lateral Sulcus
8(2-8) 9(4-10)
Frontal Horn
10(6-9) 10(7-9)
Efficiency
SIFT SURF
Cave of Septum Pellucidum
11% 11%
Superior Sagital Sinus
7% 8%
Chroid Plexus
9% 11%
Lateral Sulcus
3% 3%
Frontal Horn
50% 57%
5 CONCLUSIONS
The automatic identification of brain structures is
one of the main stages to process neuroimaging
studies. An approach to automatize it consists of
detecting landmarks over the image that features
determinate characteristics of location and intensity
values. Our research group has proposed to use
descriptors to detect these landmarks. Descriptors
are algorithms containing information relevant about
the location and intensity values of detected
landmarks. The feasibility of using descriptors with
this goal has been studied on earlier papers. So as to
obtain better results, it is needful to introduce
changes over these algorithms. In order to evaluate
and select the more adaptable algorithm to the
context of application it is essential to design an
evaluation methodology.
In this paper, a new evaluation methodology to
evaluate descriptors for neuroimaging applications is
described. The first main goal is to evaluate these
algorithms using a general test set, obtaining
parameters to quantify processing time, pairs of
homologous points between two descriptors,
stability of these methods against scaled and rotated
images and sample’s representativeness. The second
main goal is to evaluate the application of
descriptors to identify brain structures. This
evaluation will be used to select the most adequate
algorithm in a neuroimaging application context.
REFERENCES
Bay H. et al. SURF: Speeded Up Robust Features.
Computer vision and Image understanding, vol. 110,
nº 3, pp. 346-359, 2008
Gossow G. et al. An evaluation of open source SURF
implementations. RoboCup 2010, pp. 169–179, 2011.
Izard C et al. Automatic landmarking of magnetic
resonance brain images. Medical Imaging 2005:
Image Processing, vol. 5747, n. 1, pp. 1329-1340,
2005.
Lowe D. Object recognition from local scale-invariant
features. Proceedings of the International Conference
on Computer Vision, nº2, pp. 1150-1157, 1999
Lui M. L. et al. Automatic Landmark Tracking and its
Application to the Optimization of Brain Conformal
Mapping. 2006 IEEE Computer Society Conference on
Computer Vision and Pattern Recognition, vol. 2, pp.
1784 – 1792, 2006.
Luna M. et al. Automatic brain anatomical landmark
detection. Proc. Joint Workshop on New Technologies
for Computer/Robot Assisted Surgery, ISBN
9789460185496, 2012.
Mikolajczk K. et al. A performance evaluation of local
descriptors. IEEE Trans. on Pattern Analysis and
machine intelligence, 27, nº 10, pp. 1615-1630, 2005
Shattuck D. W et al. Semi-automated method for
delineation of landmarks on models of the cerebral
cortex. J. of Neuroscience Methods, vol. 178, pp. 385-
392, 2009.
Verard L et al. Fully automatic identification of AC and
PC landmarks on brain MRI using scene analysis.
IEEE Trans. on medical imaging, vol. 16, n. 5, 1997.
EvaluationMethodologyforDescriptorsinNeuroimagingStudies
117
