
Can Software Transactional Memory Make
Concurrent Programs Simple and Safe?
Ketil Malde
Institute of Marine Research, Bergen, Norway
Keywords:
Software Transactional Memory, Genome Assembly.
Abstract:
Parallel programs are key to exploiting the performance of modern computers, but traditional facilities for
synchronizing threads of execution are notoriously difficult to use correctly, especially for problems with
a non-trivial structure. Software transactional memory is a different approach to managing the complexity
of interacting threads. By eliminating locking, many of the complexities of concurrency is eliminated, and
the resulting programs are composable, and thus simplifies refactoring and other modifications. Here, we
investigate STM in the context of genome assembly, and demonstrate that a program using STM is able to
successfully parallelize the genome scaffolding process with a near linear speedup.
1 INTRODUCTION
As multi-core processors are becoming common-
place, parallel programs are crucial for performance
critical computation. Many problems can easily be
partitioned into subproblems that can be solved inde-
pendently (so-called “embarrassingly parallel” prob-
lems) but other problems are inherently more com-
plicated, and are best solved by multiple interacting
threads. In this case, care must be taken to keep sepa-
rate threads of execution from interacting in ways that
cause the program to behave incorrectly.
Traditionally, the shared data in parallel programs
is protected by synchronization primitives (locks) that
prevent simultaneous access to data structures. How-
ever, it is still quite difficult to write correct programs
using these primitives, and incorrect or careless usage
cause well-known problems like deadlocks and race
conditions (Lee, 2006). In addition, independent pro-
gram parts that use locking primitives are in general
not composable, and for instance, refactoring a pre-
viously correct program can introduce new synchro-
nization problems (Harris et al., 2005).
Software transactional memory (Shavit and
Touitou, 1995), or STM, represents a different ap-
proach. Here, state that is shared between threads
is accessed in transactions, and the state is stored in
transactional variables. If multiple threads run simul-
taneous transactions that attempt to modify the same
state, only one of the transactions succeeds, the others
are rolled back and will be rescheduled by the run-
time system.
Since there is no explicit locking, deadlocks are
eliminated, and transactions are either committed
completely or not at all, so intermediate (and possibly
inconsistent) state is never exposed. In addition, STM
transactions are composable (Harris et al., 2005). The
disadvantage is a potentially higher overhead, both
because transactions need to log access to transac-
tional variables, and because transactions sometimes
need to be restarted from scratch, which duplicates
work.
Here, we investigate how STM can be applied to
the problem of genome scaffolding, the process where
the components of a partially assembled genome se-
quence are ordered and oriented to provide a more co-
herent (but often discontiguous) whole. A scaffolder
program is implemented in Haskell using STM, and
achieves a near linear speedup with the number of
processors.
1.1 Software Transactional Memory in
Haskell
There exist implementations of software transactional
memory for many programming languages (e.g.,
Brevnov et al., 2008; Ni et al., 2008). Some of the
problems faced by implementers is that the encapsu-
lation of transactions is not easily enforced, and ex-
ceptions, I/O operations and global, mutable state can
break the transaction abstraction. Harris et al. (2005)
223
Malde K..
Can Software Transactional Memory Make Concurrent Programs Simple and Safe?.
DOI: 10.5220/0004326702230228
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2013), pages 223-228
ISBN: 978-989-8565-35-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

discuss this in more detail.
One distinguishing feature that sets Haskell apart
from the majority of programming languages, is that
it is pure: the result of a function depends only on
its parameters, and the return value may not depend
on or affect external state, read or write files, or
have other external effects. However, many effect-
ful computations can be simulated in pure code (e.g.
state can be passed between functions as a parame-
ter), and Haskell uses a structure (or pattern) called a
monad for convenient manipulation of effectful com-
putations. In essence, a monad allows the creation of
an environment where specific effects are made avail-
able. This can also include non-pure effects, and, un-
surprisingly, I/O operations are only available in the
appropriate monad.
1
The type system distinguishes effectful computa-
tions from pure computations, and enforces that pure
computations never can execute impure operations.
For instance, I/O operations are guaranteed to only
be executed in the context of the IO monad. A monad
is a parametric type, so for some type
a
, the type
IO
a
designates an I/O action which can be executed to
produce a value of type
a
. For instance,
getChar
has
type
IO Char
, as it is an I/O action that can produce
a character. Apart from the ability to be executed by
the run-time system,
getChar
is a normal value, and
like other values it can be assigned to variables and
manipulated with functions. Using combining func-
tions, larger programs can be built that interact with
their environment in complex ways.
In Haskell, STM is implemented as a monad, and
transactions are confined to this environment. Sim-
ilar to the IO example, a type
STM a
designates a
transaction that, when executed, returns a value of
some type
a
. In the STM monad, mutable data struc-
tures are available as explicitly declared transactional
variables, or
TVar
s. Using the same mechanism and
syntax as other monads, simple transactions can be
composed into more complex ones. Transactions can
be executed in the IO monad, using the
atomically
function, which converts a value of type
STM a
to a
value of type
IO a
.
It is important to note that
TVar
s are only ac-
cessible from the STM monad. This makes them
unavailable to non-transactional computations (i.e.,
plain functions), and the static type system rigidly en-
forces this encapsulation. Similarly, transactions have
no means to modify other state, in particular, they are
prevented from performing I/O operations or mod-
ifying global variables. This separation makes the
1
While most monads can be – and usually are – imple-
mented as simple libraries, the IO monad is special, and
executed by the run-time system.
Haskell STM implementation safer to use, and may
explain why STM implementationsin other languages
with less rigid type systems have been less successful.
1.2 Genome Assembly and Scaffolding
The sequencing process usually produces a large set
of short fragments (or reads) from random positions
in the genome. Given such a set of reads, the genome
assembly problem is to reconstruct the originating
genome sequence. The traditional approach is the
method called overlap–layout–consensus (Bonfield
et al., 1995; Myers et al., 2000), or OLC:
1. Identify overlaps by aligning each sequence
against all others
2. Determine the layout – order and orientation – of
the reads that is best supported by the alignments
3. Merge sequences according to layout to produce a
single contiguous consensus sequence
The first step is trivially parallelizable (each read
is independent of the others, and can be independently
aligned), but the second step is more complicated.
Usually, the problem is modeled as a graph where
each read is a node, and there exists an edge between
nodes if the corresponding reads are determined to
overlap. Assembly is then equivalent to identifying
a Hamiltonian path in the graph, which is an NP-
complete problem.
2
The layout phase processes the overlap graph to
produce a linear progression of the reads, and al-
though distant parts of the graph can be processed
independently, care must here be taken if two oper-
ations attempt to modify the same nodes simultane-
ously. The implementation details of assemblers are
not often published, but observation of some com-
mon OLC assemblers indicates that they commonly
perform alignments in parallel, but later run the lay-
out phase using a single thread of execution.
3
This
supports the view that constructing a correct locking
scheme for doing graph updates in parallel is diffi-
cult. In addition, it would probably be inefficient,
as it would incur locking overhead also for the non-
colliding updates - likely to be the vast majority of
them.
2
A popular alternative to OLC is the de Bruijn assem-
bly (Pevzner et al., 2001). This is less resource-intensive,
as it avoids the all-against alignment phase, and it is equiv-
alent to identifying an Eulerian path. But it is also easier
to parallelize in practice, which may also be a factor that
contributes to its popularity.
3
E.g. Newbler only parallelizes computing alignments
and generating output. (454 Life Sciences Corp., 2010)
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
224
Genome scaffolding is closely related to assem-
bly. Here, the assumption is that a genome has been
sequenced and assembled into a set of contigs. In
addition to overlaps, there exists external informa-
tion about the orientation and order of the contigs.
This is typically a set of paired reads, where the
members of the read pairs are separated by some ap-
proximately known distance. As for assembly, it is
not straightforward to implement a parallel scaffold-
ing algorithm correctly using locking, and commonly
used programs like SSPACE (Boetzer et al., 2011) are
single-threaded.
Scaffolding simplifies the process in two ways:
first, it reduces the amount of data that needs to be
considered (E.g. for the sea louse assembly, the ini-
tial assembly involves up to one billion reads). Sec-
ond, mapping reads to contigs make it practical to use
standard alignment tools and file formats. For these
reasons, the following will focus on the scaffolding
problem.
2 ALGORITHM AND
IMPLEMENTATION
A practical scaffolding program is likely to involve
different heuristics to resolve ambiguous cases in-
cluding repeats and chimeric contigs. As the pur-
pose here is to demonstrate STM as an implementa-
tion technique, we implement a basic scaffolding al-
gorithm that simply links together any pair of con-
tigs that has a mutual best match, as described be-
low. Matches are determined from aligned read pairs
provided as a BAM (The SAM Format Specification
Working Group, 2011) file.
First, the input BAM file is processed. By examin-
ing read pairs that map to the same contig, we obtain
estimates for the expected distance between paired
reads (called the insert length), and its variance. Also,
the total number of contigs is extracted from the BAM
file. Simultanously, the alignments relevant for scaf-
folding are extracted. In other words, each read of a
pair must map near the ends of different contigs, and
they must be oriented correctly. These alignments are
stored in an associative data structure.
The scaffolding process uses two arrays, the con-
tig array, which maps each contig to its scaffold, and
the scaffold array, which for each scaffold stores the
scaffold layout, i.e., the set of ordered and oriented
contigs. Initially, each contig is in its own singleton
scaffold.
The program now iterates over all contigs. For
each contig c, the set of read pairs with one mem-
ber matching near the 5’ end of c are extracted. The
a
b
e
c
d
C1
C2
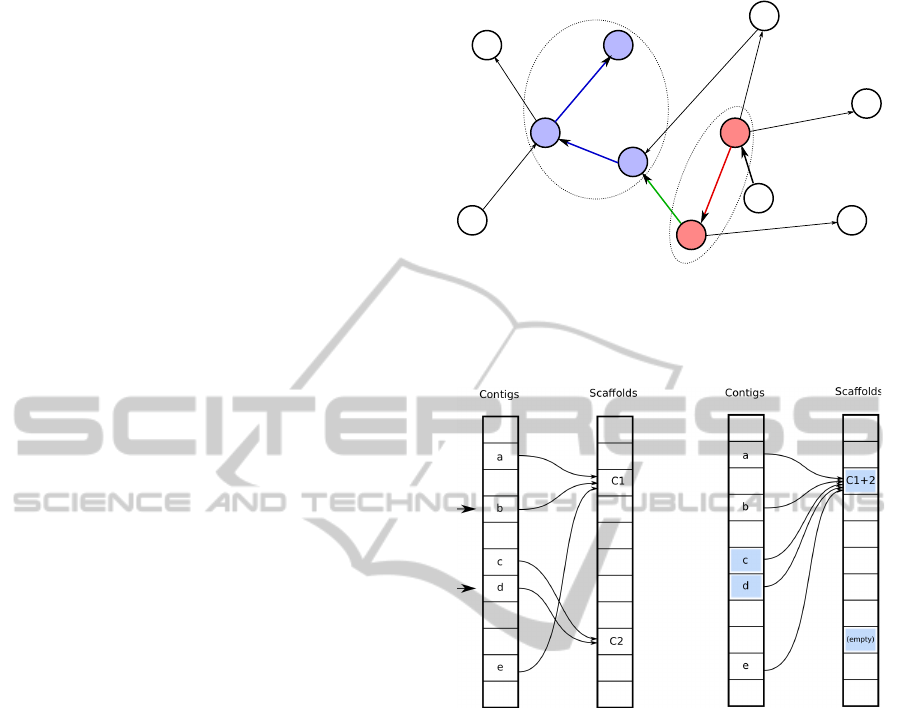
Figure 1: An example overlap graph. Two scaffolds are
already identified,
C1
(blue) containing nodes
a
,
b
, and
e
,
and
C2
(red) containing nodes
c
and
d
. Adding the edge
(green) from
d
to
b
will merge these into a single scaffold.
Contigs
Scaffolds
C1
C2
a
b
c
d
e
Contigs
Scaffolds
Figure 2: A schematic presentation of the arrays used in the
scaffolding algorithm. As in Fig. 1, contigs
a
,
b
, and
e
are initially (left) in scaffold
C1
, and contigs
c
and
d
are in
scaffold
C2
. When the algorithm decides that contigs
b
and
d
(indicated by arrows) should be adjacent, the scaffolds are
merged, causing several cells to be updated (shaded, right).
contig c
l
to which the largest number of the mapped
reads’ mates map is identified. If this relationship is
reciprocal (i.e, the reads pairs that map to c
l
have a
majority of mates mapped to c), the contigs c and c
l
are merged. The procedure is then applied similarly
to the 3’ end of c.
For instance, in the example graph in Figure 1, ex-
amination of node
b
has determined that most mapped
reads link it to
d
, and conversely, most reads mapping
to
d
link it back to
b
. This causes these two contigs
to identified as adjacent, and their scaffolds are con-
sequently merged.
Merging two scaffolds involves updating one scaf-
fold’s entry in the scaffold array to contain the new
scaffold, and deleting the other scaffold’s entry (see
Figure 2). Then, the elements in the contig array cor-
CanSoftwareTransactionalMemoryMakeConcurrentProgramsSimpleandSafe?
225

responding to contigs in the scaffold that was deleted
are updated to point to the new scaffold.
To parallelize, we simply split the iteration of
the contig array so that each thread iterates over an
equally sized segment of the array. Note that even
if threads work on separate array segments, they will
affect contigs outside their segment.
Statistically, the merging operations will usually
be independent if the arrays are large compared to
the number of concurrent operations (threads). This
also depends on the locality of merging criteria. The
current implementation considers a subgraph consist-
ing of three contigs at a time, but it could be ex-
tended to examine several candidates and links, in ef-
fect making the decision depend on a larger subset
of the graph. This would increase the chance of col-
liding operations. In any case, collisions will occur
occasionally, and a parallel implementation must take
them into account.
STM here makes this process easy, and in fact,
the code implementing this algorithm using mutable
arrays in the IO monad and using transactions in the
STM monad is exactly the same. Only the top-level
function is different, as the STM version must spawn
multiple threads that process an array segment each.
3 RESULTS
In order to test the implementation, the contigs re-
sulting from the assembly of sea louse (Lepeoph-
theirus salmonis) sequences were used. This as-
sembly was constructed using the Newbler program
(Roche), which assembled approximately 50 million
454 reads (Margulies et al., 2005) into 292421 con-
tigs.
As our pairing data, we use a set of 72 200 652 Il-
lumina reads, where each pair consists of two 100bp
reads, spaced about 150bp apart. The reads were
aligned using BWA (Li and Durbin, 2009), resulting
in 68 569 814 alignments (95% of the reads), of these
10 187580 alignments mapped the read and its mate
to different contigs.
The program was compiled with GHC 7.0.2, us-
ing the
-O2
option. It was executed on a computer
with eight Intel Xeon E7340 processors, using options
+RTS -A100M
. The parallel STM version was addi-
tionally compiled with
-threaded
, and run with
-qg
.
Figure 3 shows the running time for the scaffold-
ing stage. We see that there is some overhead associ-
ated, both with using arrays of transactional variables
(
TArray
) over regular mutable arrays (
IOArray
), and
with running on the multi-threaded GHC run-time
over the single-threaded one.
0
50
100
150
200
250
300
350
400
Array STM-S STM-1
Scaffolding time (seconds)
Figure 3: Speed spent in the scaffolding stage. “Array” is
the implementation using mutable arrays, “STM-S” is the
STM implementation running on the single-threaded run-
time, and “STM-1” is the STM implementation using a sin-
gle thread with the threaded run-time.
1
2
4
8
16
1 2 4 8 16
Scaffolding speedup
Threads
Figure 4: Speedup of the STM implementation with in-
creasing number of threads. The blue line indicates the rel-
ative performance of the non-STM (“Array”) implementa-
tion.
The STM implementation scales well. From Fig-
ure 4, we see that as we increase the number of par-
allel threads, the speedup is close to the optimum, up
to eight threads, matching the number of CPUs. The
CPUs use hyperthreading, and each processor core
appear to the OS as two processing units. Thus, the
STM implementation is still achieving a substantial
speedup going from 8 to 16 threads, even though it
means running two threads per physical core.
The resulting scaffolds were checked against scaf-
folds produced by SSPACE, and were found to differ
slightly, but for the most part, they identified the same
layout of contigs.
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
226

4 DISCUSSION AND
CONCLUSIONS
Software transactional memory is most attractive
when the program can be structured as set of mostly-
independent operations, and where each operation
only involves a small set of variables. If the oper-
ations are completely independent, the problem most
likely can be trivially partitioned, and if the number of
variables involved in each operation is large, perfor-
mance will deteriorate as the transaction log increases
in size.
The overlap-layout-consensus approach to the se-
quence assembly problem fits well with these criteria,
and is well suited to an STM approach. In the im-
plementation presented, we observe a small overhead
for using software transactional memory compared to
regular arrays, and an additional overhead for using
a multi-threaded implementation compared to a sin-
gle threaded one, but the STM implementation scales
well with the number of threads, and already with two
threads it is substantially faster. Although the results
here are very promising, it remains to be seen how far
they generalize, both as the number of CPUs increase,
and to variations of the algorithm.
This analysis has concentrated on how to improve
the run-time performance of the scaffolding process.
This is an important goal in itself, but it is even
more important to improve the quality of the result-
ing genome assembly.
The composability of STM lets the programmer
easily refactor the program or otherwise modify the
algorithm without introducing deadlocks or other syn-
chronization problems. For instance, the current
implementation only considers the potential nearest
neighbors of each contig. Extending it to take into ac-
count a larger subgraph is one possibility in improv-
ing the result. With a traditional locking scheme, this
would likely increase the complexity substantially.
With STM, it would at worst increase the chance of
collisions between transactions, leading to more re-
tries, and consequently a slightly slower program.
The source code for the implementation is avail-
able
4
under the General Public License.
REFERENCES
454 Life Sciences Corp. (2010). 454 Sequencing System
Software Manual, v 2.5p1, part C. 454 Life Sciences
Corp., Branford, CT 06405.
4
http://malde.org/∼ketil/biohaskell/stmasm
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and
Pirovano, W. (2011). Scaffolding pre-assembled con-
tigs using SSPACE. Bioinformatics, 27:578–579.
Bonfield, J. K., Smith, K. F., and Staden, R. (1995). A
new DNA sequence assembly program. Nucleic Acids
Research, 23:4992–4999.
Brevnov, E., Dolgov, Y., Kuznetsov, B., Yershov, D.,
Shakin, V., Chen, D.-Y., Menon, V., and Srinivas, S.
(2008). Practical experiences with java software trans-
actional memory. In Proceedings of the 13th ACM
SIGPLAN Symposium on Principles and practice of
parallel programming, PPoPP ’08, pages 287–288,
New York, NY, USA. ACM.
Harris, T., Marlow, S., Peyton-Jones, S., and Herlihy,
M. (2005). Composable memory transactions. In
Proceedings of the tenth ACM SIGPLAN symposium
on Principles and practice of parallel programming,
PPoPP ’05, pages 48–60, New York, NY, USA. ACM.
Lee, E. A. (2006). The problem with threads. Technical
Report UCB/EECS-2006-1, EECS Department, Uni-
versity of California, Berkeley. The published version
of this paper is in IEEE Computer 39(5):33-42, May
2006.
Li, H. and Durbin, R. (2009). Fast and accurate short read
alignment with burrows-wheeler transform. Bioinfor-
matics, 25:1754–1760.
Margulies, M., Egholm, M., Altman, W. E., Attiya, S.,
Bader, J. S., et al. (2005). Genome sequencing in mi-
crofabricated high-density picolitre reactors. Nature,
437:376–380.
Myers, E. W., Sutton, G. G., Delcher, A. L., Dew, I. M.,
Fasulo, D. P., Flanigan, M. J., et al. (2000). A
whole-genome assembly of drosophila. Science,
287(5461):2196–2204.
Ni, Y., Welc, A., Adl-Tabatabai, A.-R., Bach, M.,
Berkowits, S., Cownie, J., Geva, R., Kozhukow, S.,
Narayanaswamy, R., Olivier, J., Preis, S., Saha, B.,
Tal, A., and Tian, X. (2008). Design and implementa-
tion of transactional constructs for c/c++. In Proceed-
ings of the 23rd ACM SIGPLAN conference on Object-
oriented programming systems languages and appli-
cations, OOPSLA ’08, pages 195–212, New York,
NY, USA. ACM.
Pevzner, P. A., Tang, H., and Waterman, M. S. (2001).
An eulerian path approach to dna fragment assem-
bly. Proceedings of the National Academy of Sciences,
98(17):9748–9753.
Shavit, N. and Touitou, D. (1995). Software transactional
memory. In Proceedings of the fourteenth annual
ACM symposium on Principles of distributed comput-
ing, PODC ’95, pages 204–213, New York, NY, USA.
ACM.
The SAM Format Specification Working Group (2011). The
SAM Format Specification.
APPENDIX
The code for the merging operation (as illustrated in
Figure 2 is given below. Note that the type signature is
CanSoftwareTransactionalMemoryMakeConcurrentProgramsSimpleandSafe?
227

not given, and the code will typecheck and run with-
out modification in either the IO monad or the STM
monad. In STM, each array cell is a TVar, and the
merge operation must be part of a transaction. If an-
other thread modifies any of the involved array loca-
tions before the transaction completes (say by merg-
ing one of the clusters with a different cluster), the
transaction will be aborted and restarted. In IO, there
are no such guarantees, and the function can only be
run safely in a single thread.
The function takes as input parameters arrays of
contigs (each pointing to a cluster) and scaffolds
(echo containing the list of its elements), and a pair
of contigs. It then merges the scaffolds that contain
the given each contig from the pair.
merge contigs scaffolds (contig1,contig2) = do
-- Get the scaffolds for each contig
i1 <- readArray contigs contig1
i2 <- readArray contigs contig2
when(i1/=i2) $ do
-- read counts and elements from clusters
(n1,cs1) <- readArray scaffolds i1
(n2,cs2) <- readArray scaffolds i2
-- write the merged cluster in i1,
-- and an empty cluster in i2
writeArray scaffolds i1 (n1+n2,cs1++cs2)
writeArray scaffolds i2 (0,[])
-- update previous elements in i2
-- to point to the merged cluster
mapM_ (\x -> writeArray contigs x i1) cs2
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
228
