
Finger Joint Characterization from X-Ray Images for Rheumatoid
Arthritis Assessment
Joan M. N´u˜nez, D´ebora Gil and Fernando Vilari˜no
Dept. of Comp. Science, Comp. Vision Center, Edifici O - UAB, 08193 Bellaterra, Spain
Keywords:
Rheumatoid Arthritis, X-Ray, Hand Joint, Sclerosis, Sharp Van der Heijde.
Abstract:
In this study we propose a modular system for automatic rheumatoid arthritis assessment which provides a joint
space width measure. A hand joint model is proposed based on the accurate analysis of a X-ray finger joint
image sample set. This model shows that the sclerosis and the lower bone are the main necessary features in
order to perform a proper finger joint characterization. We propose sclerosis and lower bone detection methods
as well as the experimental setup necessary for its performance assessment. Our characterization is used to
propose and compute a joint space width score which is shown to be related to the different degrees of arthritis.
This assertion is verified by comparing our proposed score with Sharp Van der Heijde score, confirming that
the lower our score is the more advanced is the patient affection.
1 INTRODUCTION
Rheumatoid arthritis (RA) is a chronic disease that
causes joint dysfunction which results, among other
symptoms, in the reduction of the joint space width
(JSW) and the erosion of the joint bones. Since there
is not any cure for RA, the assessment of the disease
becomes very important and it has led to the emer-
gence of several semi-quantitative assessment meth-
ods. Among them, the Sharp-Van der Heijde (SvdH)
score, which provides separate discrete values for
JSW and bone erosion based on X-ray image exam-
ination, is the most widely used nowadays (Van der
Heijde, 1999). The patient score is the sum of the
scores for all the considered joints in hands and feet.
Three stages should be considered to face the au-
tomation of RA assessment: joint detection, joint
characterization and joint measurements. As the main
goal of this study is to propose and assess a JSW mea-
sure we will focus on the second and third stages tak-
ing hand joints as input (Figure 1). This statement
leads us to a hand joint modelling to describe the nec-
essary features to compute JSW measures. Sclerosis
and lower bone appear to be the main necessary fea-
tures to obtain JSW measures and the corresponding
detectors are proposed.
Few previous works have faced this problem. Bi-
elecki et al. (Bielecki et al., 2008) developed the first
study that faced the challenge of a fully automatic sys-
tem for RA assessment. Vera et al. (Vera, 2010) pro-
vided a method which included all the different stages
of the problem improving significantly the joint de-
tection rates. Langs et al. (Langs et al., 2009) pre-
sented a new method whose scope is also the whole
RA assessment process. However, these works faced
the problem as a whole stage, not modular, and did
not show a clear correlation among its proposed JSW
score and Svdh.
Our proposal clearly separates joint detection,
joint characterization and joint measurements in three
independent modules. It also provides a framework
for sclerosis and lower bone detection performance
assessment, and proposes a JSW score which exhibits
a close relation with SvdH score. This approach al-
lows us to confirm for the first time a relation between
an automatic score and the RA disease stage. More-
over, the modular orientation of our contribution, un-
like previous approaches, enables future research to
specifically focus on the improvement of the different
open challenges independently.
2 DATA ANALYSIS
This study is focused on the same digital database
used by Vera et al. (Vera, 2010) enriched with our
own manually segmented ground truth for the se-
lected visual features necessary for JSW computation.
Twenty X-ray images from different patients contain-
ing pairs of hands in posterioanterior view are avail-
288
Núñez J., Gil D. and Vilariño F..
Finger Joint Characterization from X-Ray Images for Rheumatoid Arthritis Assessment.
DOI: 10.5220/0004327302880292
In Proceedings of the International Conference on Biomedical Electronics and Devices (BIODEVICES-2013), pages 288-292
ISBN: 978-989-8565-34-1
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
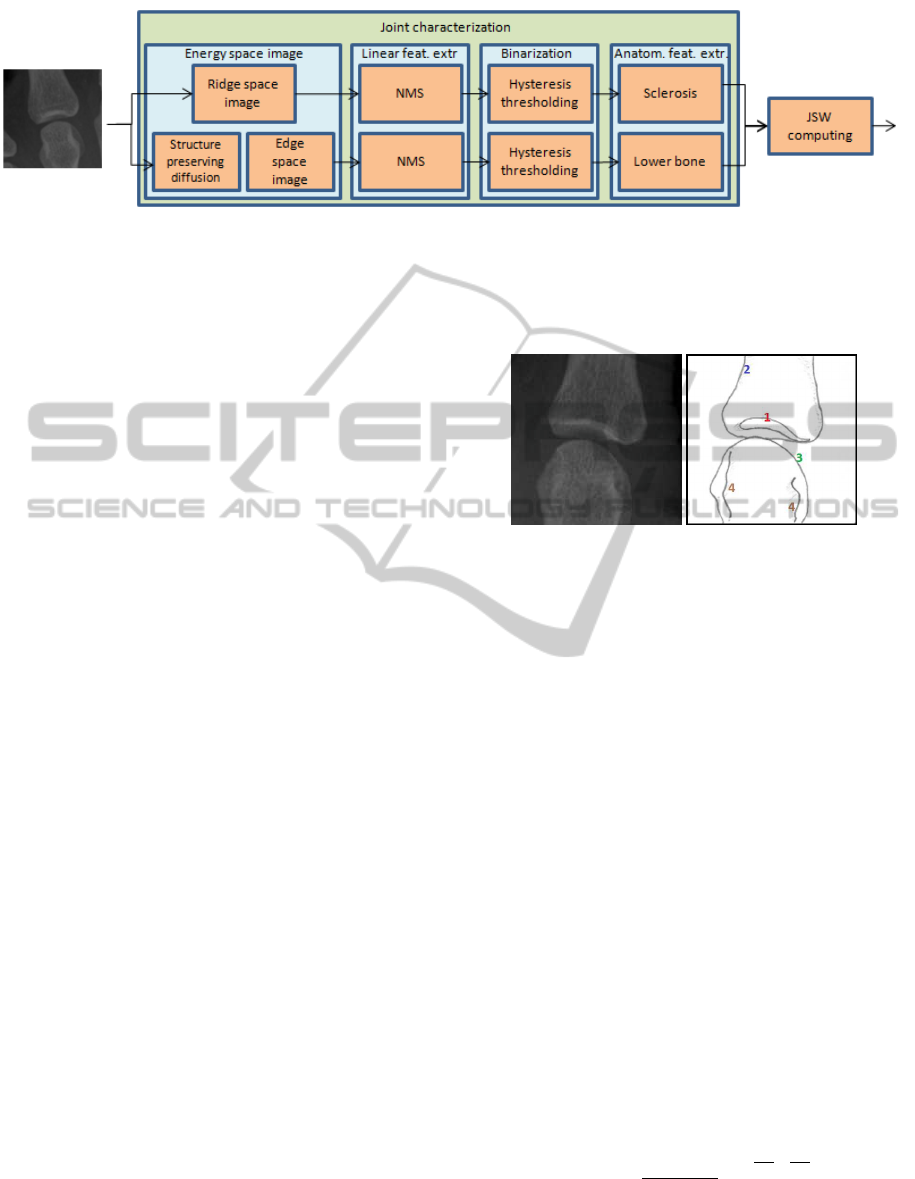
Figure 1: Rheumatoid Arthritis characterization system.
able in DICOM format, 2828x2320 resolution and in-
tensity range from 0 to 4095. Hand joints have been
marked by an expert who was asked to spot the joint
middle point along the finger axis and the joint ori-
entation. A hand joint dataset containing 560 images
of joints have been created (14 joints/hand). Among
these samples, a total number of 320 joints were
labelled with their SvdH score for inter-phalangeal
distance (discrete values from 0 to 3). These 320
samples comprised 160 metacarpo-phalangeal joints
(MCP) and 160 proximal inter-phalangeal joints (PIP)
-distal inter-phalangeal joints (DIP) are not consid-
ered within SvdH score-. Erosion scoring was not col-
lected because it is out of the scope of this this study.
Our proposed hand joint model does not consider
thumb joints as they represent a different problem due
to its specific profile. Since images are taken with the
hands in posterioanterior view -from the hand back
with the palm facing down- a profile view of the
thumb is taken. Consequently, a frontal projection
is obtained for the rest of the fingers whereas a lat-
eral projection is obtained for the thumbs. This rea-
son, as well as their specific skeletal structure, causes
the visual features observed in thumbs to be substan-
tially different and justifies that they are not included
in this study. As far as the other finger joints are
concerned, several features are distinguished within
the model: sclerosis, upper bone contour, lower bone
contour and lower bone inner edges, as depicted in
Figure 2 (N´u˜nez et al., 2011). The sclerosis and the
lower bone contour are the main necessary features to
carry out measures on JSW. The sclerosis (feature 1
in Figure 2(b)) appears as a consequence of the up-
per bone shape and the way an X-ray image is cre-
ated. The X-ray beam have to pass through a higher
density region due to the concavity of the lower part
of the upper bone of the joint (feature 2 in Figure
2(b)). As a consequence, the sclerosis appears as a
prominent high intensity region. Regarding the lower
bone, the visual features are also a consequence of the
mentioned X-ray image acquisition mechanism and
the lower bone shape (feature 3 in Figure 2(b)). That
shape can vary in the different fingers or the different
hand joints and the inner edges may be present or not
(feature 4 in Figure 2(b)). However, the lower bone
contour is defined by the outer part of the lower bone.
(a) (b)
Figure 2: (a) Joint example. (b) Joint model schema (1:
sclerosis, 2: upper bone, 3: lower bone, 4: inner edges).
3 METHODOLOGY
Our proposed sclerosis and lower bone detectors in-
clude several stages: energy space image, linear fea-
ture extraction, binarization and anatomical feature
extraction.
The first stage aims to obtain the energy space im-
age. The intensity levels of that image describe the
chances of finding a feature in every pixel. Two sepa-
rate processes are achieved in order to obtain the ridge
space image and the edge space image. A second
derivative of anisotropic Gaussian (DoG) method is
used to compute the ridge space image. This method
is based on the fact that ridges can be obtained as
the high values in the second derivative. We apply a
bank of filters where each of them achieves a convo-
lution with the corresponding kernel template. We de-
sign our filter templates based on second derivatives
of anisotropic Gaussian kernels. Considering that an
oriented Gaussian function is described by:
G
(σ
x
,σ
y
),θ
=
1
(2π)σ
x
σ
y
e
−
˜x
2
2σ
2
x
+
˜y
2
2σ
2
y
(1)
where (σ
x
, σ
y
) are the scales in the corresponding
axis, θ is the rotation angle of the filter and ˜x and ˜y
FingerJointCharacterizationfromX-RayImagesforRheumatoidArthritisAssessment
289

are the coordinates given by the rotation angle. Hence
they are defined as:
˜x = xcosθ+ ysinθ
˜y = xsinθ − ycosθ
(2)
We use anisotropic Gaussians with σ = σ
x
= 2σ
y
.
Therefore, the Gaussian function results in:
G
σ,θ
=
1
(2π)2σ
2
e
−
˜x
2
2(2σ)
2
+
˜y
2
2σ
2
(3)
Hence the kernel will be defined as:
∂
2
˜y
G
σ,θ
=
˜y
2
− 1
σ
4
G
σ,θ
(4)
We apply a normalization so that the geometry of
the valleys is prioritized:
G
N
σ,θ
:=
k∂
2
˜y
G
σ,θ
∗ Ik
k∂
2
˜y
G
σ,θ
kkIk
(5)
where k · k stands for the L
2
integral norm and ∗ de-
noting the convolution operator.
The kernels are applied to different scales and in
different equally distributed orientations in a range
centered in the joint orientation. The final outcome
are 24 output images, each of them corresponding to
a determined orientation and scale. Hence, the output
I
ridges
must be a combination of all of them, defined
as the maximum of the outputs from each filter:
I
ridges
= max
i, j
G
N
σ
i
,θ
j
(6)
On the other hand, the edge space image is ob-
tained as the gradient of the input image after apply-
ing structure preserving diffusion (Gil et al., 2009).
Diffusion filtering proved its success in improving the
quality of the edge detection by smoothing the image
irregularities while keeping the main image structure.
The second stage performs the feature extraction
using Non-maximum Suppresion algorithm (Canny,
1986), which only keeps pixels that are local maxima
along the gradient direction. Gradients are computed
using the structure tensor of the space image.
In the third stage hysteresis thresholding algo-
rithm (Canny, 1986) is used to binarize the non-
maximum suppressed images while preserving fea-
ture connectivity and removing weak responses.
Finally, the two images forwarded by the third
stage must go through the fourth processing step in or-
der to provide the final sclerosis and lower bone seg-
mentations. This stage is different for the two thresh-
olded images:
• The final sclerosis segmentation is the ridge in the
binarized image that is closer to the center of the
image following the finger orientation .
• As far as lower bone is concerned, the correspond-
ing binarized image is processed to remove the
edges in the upper part and the margins. After-
wards, the endpoints in the remaining processed
edges are linked. The final lower bone segmenta-
tion is obtained by computing the convex hull of
the linked-edge image.
4 EXPERIMENTAL SETUP
Two subsets were created from our dataset of 320 an-
notated images to perform our experiments: 1) Tune
Dataset, with 40 randomly selected healthy joints (20
MCP and 20 PIP); 2) Test Dataset, with the remaining
280 joints (140 MCP and 140 PIP). The Tune Dataset
was used to tune the parameters of the system for scle-
rosis and lower bone contour segmentation, and the
Test Dataset was used to compute the output of our
system and compare the proposed distance measure
with the SvdH score. We added 20 DIP joints from
the non-annotated dataset to enrich the variability of
the Tune Dataset -these joints can be safely included
because, although not having a SvdH score, they fit
our model-. Both sclerosis and lower bone were man-
ually segmented by an expert using OsiriX (Rosset
et al., 2004) software exclusively for the 60 images of
the Tune Dataset.
Performance metrics were developed to evaluate
the performance of the system, i.e. the quality of our
detections, and tune the parameters. We based our
metrics on the Average Surface Distance (ASD), de-
fined as follows:
ASD(U, V) =
1
|S(U)|
∑
s
U
∈S(U)
d(s
U
, S(V))
!
(7)
where, given a pixel p and a region R conformed by a
set of pixels S(R), d(p, S(R)) is defined as:
d(p, S(R)) = min
s
R
∈S(R)
kp − S
R
k (8)
where k.k stands for the Euclidean distance.
Thus, if A denotes our automatic segmentation and
M denotes the manual delineation, we define:
Caught = ASD(A, M) (9)
Missed = ASD(M, A) (10)
When both Caught and Missed metrics are zero
the segmentation is perfect. Caught metric value is
related to the quality of the detector at detecting valid
pixels (true positives) and avoiding non-desired pix-
els (false positives). Analogously, the lower the value
of Missed, the less desired information was missed
(false negatives).
BIODEVICES2013-InternationalConferenceonBiomedicalElectronicsandDevices
290
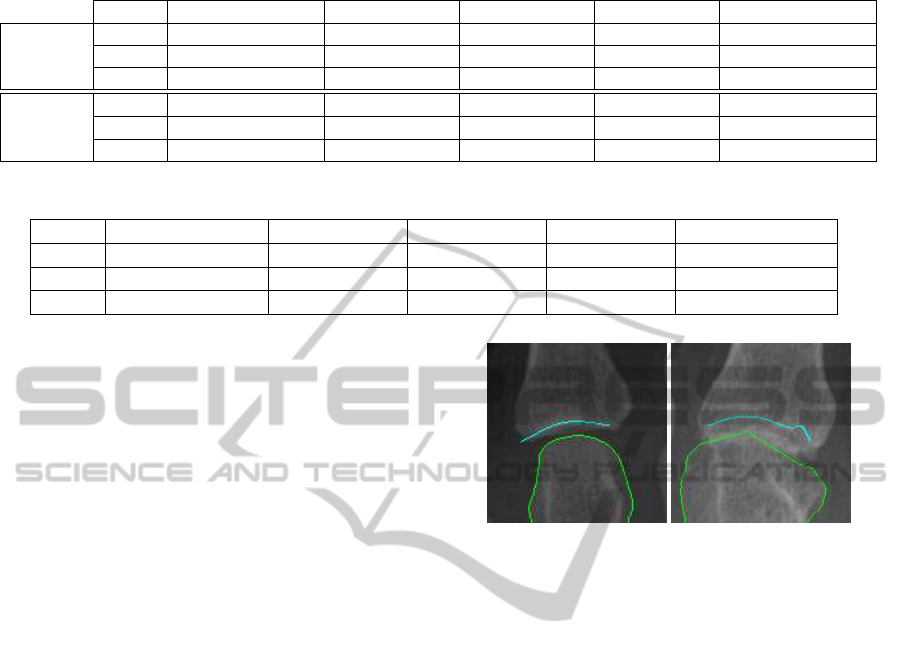
Table 1: Valid sclerosis and lower bone detection rates.
J. type SvdH 0 SvdH 1 SvdH 2 SvdH 3 Total
Sclerosis
MCP 111/115 (96.5%) 10/11 (90.9%) 10/12 (83.3%) 2/2 (100.0%) 133/140 (95.0%)
PIP 64/67 (95.5%) 27/35 (77.1%) 22/28 (78.5%) 5/10 (50.0%) 118/140 (84.2%)
Total 175/182 (96.1%) 37/46 (80.4%) 32/40 (80.0%) 7/12 (58.3%) 251/280 (89.4%)
L. bone
MCP 85/115 (73.9%) 5/11 (45.4%) 8/12 (66.6%) 0/2 (0.0%) 98/140 (70.0%)
PIP 42/67 (62.6%) 14/35 (40.0%) 8/28 (28.5%) 3/10 (30.0%) 67/140 (47.8%)
Total 127/182 (69.7%) 19/46 (41.3%) 16/40 (40.0%) 3/12 (25.0%) 165/280 (58.9%)
Table 2: Valid sclerosis and lower bone detection rates (both in the same image).
J. type SvdH 0 SvdH 1 SvdH 2 SvdH 3 Total
MCP 83/115 (72.1%) 5/11 (45.4%) 6/12 (50.0%) 0/2 (0.0%) 94/140 (67.1%)
PIP 39/67 (58.2%) 11/35 (31.4%) 6/28 (21.4%) 2/10 (20.0%) 58/140 (41.4%)
Total 122/182 (67.0%) 16/46 (34.7%) 12/40 (30.0%) 2/12 (16.6%) 152/280 (54.2%)
This experimental setup was used in order to train
the detection systems with the Tune Dataset. The
system providing the lowest values for the perfor-
mance metrics was selected, giving priority to a lower
Caught value so that the provided output is better al-
though some parts may be missed. Afterwards, the
selected detectors were applied to the Test Dataset.
The final sclerosis and lower bone outputs were then
visually tested in order to separate the wrong detec-
tions from those detections that can be useful to com-
pute JSW scores. The final detections were accepted
if they provided a good segmentation and there were
not remaining spurs in the joint interspace.
Finally, the proposed JSW measurement is the
minimum distance between the sclerosis and the
lower bone. It is defined as:
d
min
= min
p∈L
d(p, S) (11)
where S and L stand for the sclerosis surface and the
lower bone contour, respectively.
5 RESULTS
Table 1 shows the valid sclerosis and lower bone
rates when applying our sclerosis detector to the Test
Dataset. High detection rates have been obtained in
the case of sclerosis detection, reaching higher rates
in the case of healthy samples (SvdH 0), providing
96.1%, since they are closer to our joint model. How-
ever, these detection rates drop down to 58.3% in the
case of joints in a more advanced RA stage, which
makes clear the potential usefulness of the model in
order to classify advanced stages of the disease. All
in all, the detection rate reaches the remarkable value
of 89.6% on the whole dataset. As far as MCP and
PIP are concerned, the detection rates remain close
(a) (b)
Figure 3: (a) MCP valid detection. (b) MCP wrong detec-
tion.
although a light tendency is confirmed which shows
that PIP joints present more detection problems.
On the other hand, lower bone detection arises as
a very difficult problem. The overall detection rate
drops to 58.9% although the healthy samples reach
69.7%. The lack of an stardard practice in X-ray im-
age techniques causes an important variability in the
samples which makes lower bone segmentation diffi-
cult to achieve. Figure 3 shows two detection exam-
ples of wrong and valid segmentations.
The samples where both lower bone and sclerosis
segmentations were labelled as valid were considered
for further analysis. Table 2 contains the correspond-
ing results. The rates when the two segmentations are
considered altogether are just slightly below the lower
bone detection rates, reaching 67.0% for healthy sam-
ples and 54.2% for the whole dataset.
Afterwards, the proposed JSW measure was com-
puted and tested in relation to SvdH score. In the
case of MCP samples (Figure 4), the JSW estimation
shows a light decreasing trend as the value of SvdH
score increases. Nevertheless, that decreasing trend
is clear in the case of PIP samples (Figure 5). It is an
encouraging result taking into account that our system
is only considering JSW values. Standard clinical as-
sessment considers more information as the symme-
FingerJointCharacterizationfromX-RayImagesforRheumatoidArthritisAssessment
291
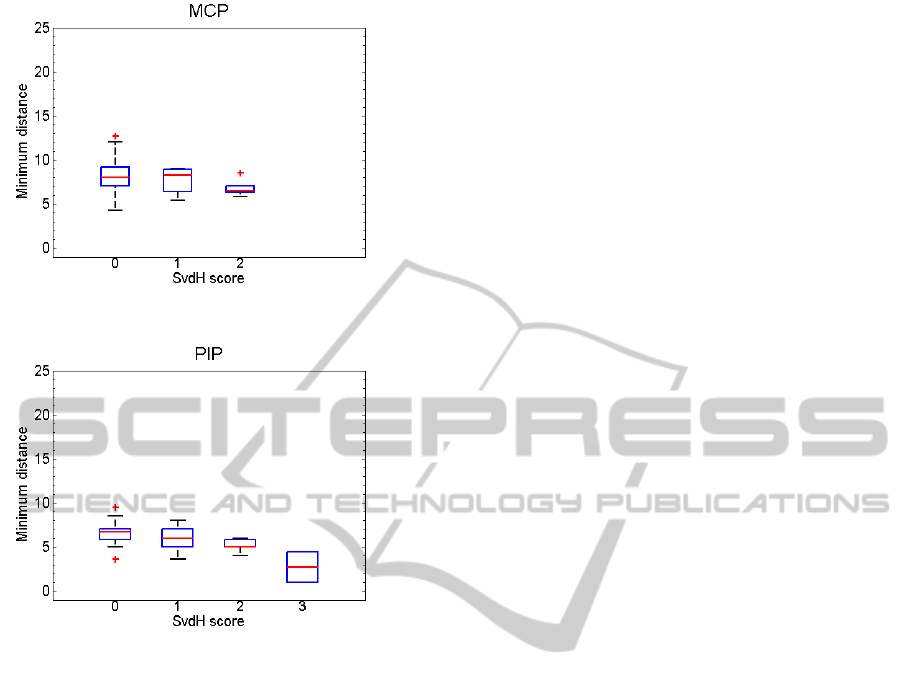
Figure 4: MCP joints, distance measure and SvdH score.
Figure 5: PIP joints, distance measure and SvdH score.
try between hand joints or erosion information.
Finally, the reliability of the proposed system to
provide a proper JSW estimation independently of its
relation to the RA stage was tested. The JSW measure
was computed both for the manual and automatic seg-
mentations of the Tune Dataset, providing a final error
of 1± 0.7 pixels. This result confirms the robustness
of the JSW estimation.
6 CONCLUSIONS
The proposed system sets the foundations of a mod-
ularized RA assessment system. We created a hand
joint sample dataset and introduced a hand joint
model based on the skeletal structure of hand bones.
Our sclerosis detector achieves remarkably good re-
sults. However, lower bone segmentation appears to
be a harder task when faced in a local way. The
JSW measure we propose was compared with the gold
standard score for RA assessment. We showed for the
first time that an automatic measure for JSW can be
computed so that its value has a clear relation to the
SvdH manual measure assessed by clinical person-
nel and, therefore, to RA disease stage. Future work
should involve the confirmation of this trend with a
dataset which should include a larger study with sam-
ples from multiple sources. Higher SvdH score sam-
ples, particularly scarce in number should be also con-
sidered.
ACKNOWLEDGEMENTS
This work was supported in part by the Spanish Gov.
grants TIN2012-33116, TIN2009-13618, MICINN
TIN2009-10435 and Consolider 2010 MIPRCV
(CSD2007-00018), and the UAB grant 471-01-
2/2010. The second author has been supported by the
Ramon y Cajal Program.
REFERENCES
Bielecki, A., Korkosz, M., and Zielinski, B. (2008). Hand
radiographs preprocessing, image representation in
the finger regions and joint space width measurements
for image interpretation. PRn, 41(12):3786–3798.
Canny, J. (1986). A computational approach to edge detec-
tion. PAMI, (6):679–698.
Gil, D., Hern`andez-Sabat´e, A., Burnat, M., Jansen, S.,
and Mart´ınez-Villalta, J. (2009). Structure-preserving
smoothing of biomedical images. In Computer Anal-
ysis of Images and Patterns, pages 427–434. Springer.
Langs, G., Peloschek, P., Bischof, H., and Kainberger, F.
(2009). Automatic quantification of joint space nar-
rowing and erosions in rheumatoid arthritis. Medical
Imaging, IEEE Transactions on, 28(1):151–164.
N´u˜nez, J., F., V., and D., G. (2011). Computer vision tech-
niques for characterization of finger joints in x-ray im-
ages. Technical report, Centre de Visi´o per Computa-
dor.
Rosset, A., Spadola, L., and Ratib, O. (2004). Osirix: An
open-source software for navigating in multidimen-
sional dicom images. Journal of Digital Imaging,
17:205–216. 10.1007/s10278-004-1014-6.
Van der Heijde, D. (1999). How to read radiographs accord-
ing to the sharp/van der heijde radiological assessment
in rheumatoid arthritis in long term studies. Journal
of Rheumatology, (26):743–745.
Vera, S. (2010). Finger joint modelling from hand x-ray
images for assessing rheumatoid arthritis. Technical
report, Centre de Visi´o per Computador.
BIODEVICES2013-InternationalConferenceonBiomedicalElectronicsandDevices
292
