
Semantic Integration for Model-based Life Science Applications
Tomasz Gubała
1,4
, Katarzyna Prymula
3
, Piotr Nowakowski
1
and Marian Bubak
2,4
1
Distributed Computing Environments Team, Academic Computer Centre Cyfronet AGH,
Nawojki 11, 30-950 Krakow, Poland
2
AGH University of Science and Technology, Department of Computer Science, Mickiewicza 30, 30-059 Krakow, Poland
3
Department of Bioinformatics and Telemedicine, Medical College, Jagiellonian University,
Sw. Łazarza 16, 31-530 Krakow, Poland
4
Informatics Institute, University of Amsterdam, Science Park 904, 1098 XH Amsterdam, The Netherlands
Keywords:
Semantic Modeling, Database Semantics, Genetics Models, In-silico Experimentation.
Abstract:
When delivering tools and solutions for modern e-Science applications, the proper transfer of domain knowl-
edge into information models is a crucial design step. In this work we describe the Semantic Integration
approach to modeling and transcribing complex science domain knowledge into well-structured information
models based on semantics. Apart from a conceptual study and presentation of related work, we also describe
how we applied the methodology to deliver a real-life solution for simulation of locations of active binding
sites in proteins – an important problem in bioinformatics. Our goal is to show how the Semantic Integration
technology can help deliver ready-to-use solutions for scientists.
1 INTRODUCTION
During the last decade we have witnessed the advent
of a new approach to scientific investigations using
computational facilities, based not only on large par-
allel supercomputers, but also employing application
servers, distributed systems and local workstations.
Researchers in areas such as modeling and simula-
tion (Guru et al., 2009) or computer-aided health-
care (Sloot et al., 2006) have been adopting a new
paradigm involving multiple steps, often performed
ad-hoc on local resources, with scripts, remote web
tools and database access helpers. The reason behind
this shift is twofold. First, it is about automation –
numerous preprocessing and output parsing and anal-
ysis tasks are becoming more and more automated to
streamline research work and increase effectiveness,
eliminating unnecessary effort and reducing the po-
tential for human error. Secondly, the ongoing devel-
opment of more agile and type-as-you-go program-
ming tools (including advanced integration mecha-
nisms for scientific workflows) has made it easier
for cross-domain educated scientists to create custom
tools for their daily research work. Automation and
dynamic experimentation also support another novel
scientific short-circuit technique – broad exploration
of solution spaces, where uninteresting avenues of re-
search can be automatically filtered out.
This gradual rise in the effectiveness of scientific
experiments performed using computational software
and hardware comes at the expense of having to main-
tain a growing body of hand-tailored scripts, parsers
and analyzers, many of which are intended as one-off
helpers for a single purpose but later tend to become
(through constant fixing and adjusting) very popular,
though convoluted, tools for a certain domain. Our
discussions with chemists and biologists indicate that
they spend a lot of their time digging through fora and
webpages for useful scraps of Python or Perl scripts.
While the very existence of such scripts (sometimes in
abundance) is a success in itself, further steps need to
be taken in terms of integration, standardizations and
interoperability. Research and deployment programs
like ESFRI
1
and EGI
23
(Bubak et al., 2012) are meant
to pave the way to integrated environments for future
system-level science.
A crucial part of our research on the development
of methods and tools for system-level science focuses
on the problem of integration and maintenance of
(sometimes quickly evolving) computational science
tools so that they may act as useful, natural toolkits for
1
European Strategy Forum on Research Infrastructures,
http://ec.europa.eu/research/infrastructure/
2
European Grid Infrastructure, http://www.egi.eu/
3
PL-Grid Project, http://www.plgrid.pl/
74
Gubała T., Prymula K., Nowakowski P. and Bubak M..
Semantic Integration for Model-based Life Science Applications.
DOI: 10.5220/0004426400740081
In Proceedings of the 3rd International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH-2013),
pages 74-81
ISBN: 978-989-8565-69-3
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

scientists. We closely observe the difficulties faced by
researchers who try to conduct complex e-science ex-
periments. From these observations we define the fol-
lowing challenge: what methodology a scientific pro-
grammer of in silico experiments should employ to in-
tegrate old and implement new software according to
the user expectations while avoiding one-off software
cludges (and thus worsening the situation for future
researchers)?
In the following sections we present an idea of se-
mantic integration (Gubala et al., 2009) which allows
scientific programmers and computational scientists
to integrate their workbench tools based on objects
and concepts from their own domain. The flexibil-
ity of the method allows for constant improvements
while still maintaining a well-defined semantic data
model that guarantees future maintainability and ex-
tensibility. Through an extensive case study of a spe-
cific application from the protein analysis domain we
prove that our proposal represents a viable solution to
the stated problem.
In order to position our work vis a vis current re-
search on web ontologies, shared taxonomies and on-
tology languages, we would like to elaborate on our
understanding of the crucial concept of ”semantics”.
In this paper, we define semantics as an assignment
of meaning to terms in the scope of a roughly de-
fined domain. In other words, we consider seman-
tics to represent an answer to question: ”What does
this term mean to you?”, asked of a domain expert
(in line with Petrie (Petrie, 2009)). We do not equate
the semantic model with an ontological description,
nor do we use ontological languages directly in our
work. By doing that, however, we do not wish to
imply that ontological modeling is in any way infe-
rior to the taxonomy-based approach we use. We also
do not claim that ontologies are not applicable in our
work – merely that their application (or lack thereof)
is not crucial from the point of view of the seman-
tic integration approach proposed. While we prefer
to use non-ontological taxonomy modeling in our ap-
plication, we remain fully aware that using ontologies
(transcribed e.g. in OWL-DL) is also possible.
2 RELATED WORK ON
SEMANTICS USED
FOR INTEGRATION
Using domain semantics for integration of subsys-
tems is an approach often applied in (scientific) work-
flow research. Since building workflows to model
data or control the flow of an application is clearly
a method of integration, it can be based on domain
knowledge in the form of ontological annotations
(Corcho et al., 2006) or present in the workflow de-
scription itself (Bubak and Unger, 2007) (Kryza et al.,
2007). We argue, however, that using complex work-
flow management and, intrinsically complex, onto-
logical modeling, might not be an option for smaller,
ad-hoc e-Science tools targeted by our research.
Substantial attention was also devoted to the in-
tegration of application assets, namely data sources
and computation elements. The variability and het-
erogeneity of data sources (in the form of databases or
medical instruments) may be tackled by applying se-
mantics as a single, common, sound data model (Can-
nataro et al., 2007). Apart from semantic annotation
of data sources, we may also perform semantic-based
dynamic query augmentation on such data, to deliver
an integrated database-like store (Kondylakis et al.,
2007). An example of semantic description of compu-
tation elements which act on data, usually to analyze
and transform it for scientific purposes, is Semantic
MOBY (P.M. Gordon, 2007). It attaches semantic in-
formation to a service so that users are able to quickly
find and use the provided tools.
Both semantic integration of data (which models
the static part of an application) and semantic annota-
tion of services (to represent the dynamic aspect of an
application as a process) could be treated as parts of
models within the solution we propose (either the do-
main or the integration model). It is therefore possible
to combine such techniques with the idea of concept
and facet separation, presented above.
3 SEMANTIC INTEGRATION
IDEA
The core idea of the proposed methodology is to es-
tablish a firm basis of the integration process in a se-
mantic, domain-specific data model and to evolution-
arily extend it with an orthogonal integration model
as new tools and processes are integrated. This de-
composition into two different submodels, being the
core element of the proposed method, helps achieve
separation of concerns for each new application (re-
quirements of the domain against requirements of the
platform). If the domain expert (i.e. the researcher)
and the developer are different persons, the method-
ology assumes relatively tight collaboration between
the two – just like in any successful computational sci-
ence project. Starting with a set of elements of the sci-
entific experiment process, such as scripts, HPC ap-
plications and remote web applications, the program-
mer first establishes the domain model.
SemanticIntegrationforModel-basedLifeScienceApplications
75

3.1 Modeling of Application Domain
The need for a domain model as a separate, indepen-
dent element, might not be so obvious at first. When
approaching a typical problem of implementing and
conducting an in-silico experiment, one usually deals
with three to four tools which should be integrated
(perhaps as a workflow). It might be enough to estab-
lish a communication protocol based on implicit data
structures between these tools to make them work to-
gether; however there are two important factors to
consider. First of all, the developer may quickly dis-
cover additional elements which have to be integrated,
e.g. a persistence DB, a new web interface with its
corresponding server and client sides, perhaps a third-
party support library etc. Hence, as a result, the fi-
nal application may have to incorporate not 3-4 but 4-
7 elements with nontrivial connections (for instance,
several domain tools may need to present something
using the web interface or may require some shared
persistence layer). Thus, the dedicated protocol ap-
proach becomes more challenging and results in less
code reuse.
Even more important is the fact that sooner or later
the tool may need to be extended, which is especially
true in modern life sciences. Therefore, expressing
the model implicitly in the protocols may severely im-
pact the maintainability and extensibility of the result-
ing solution.
For our domain model we propose a well-known
object-oriented approach. While some authors apply
ontologies to semantic domain modeling (Rodriguez
and Egenhofer, 2003), we have learned from our pre-
vious studies (Gubala and Hoheisel, 2006) that such
an approach is better suited for applications which
are very complex but also relatively stable (e.g. cri-
sis management systems for flood control or pollution
monitoring). In situations where one deployment of a
tool serves for, at most, several months (sometimes
weeks) and then needs to be revised, ontology-based
taxonomies are too complex and not flexible enough
to be an effective modeling solution. While ontology-
based description logic has its advantages over an ob-
ject model, we do not consider these as necessary to
properly model domain semantics. Another important
factor taken into consideration was the fact that pro-
grammers are far more familiar with object modeling
techniques (as they are part of any typical computer
science bachelor’s curriculum) compared to ontolog-
ical modeling – this relatively lenient learning curve
translates into better adaptability for recently formed
e-science teams.
Figure 1 shows a small part of our domain model
constructed for the HIV drug resistance domain in the
Figure 1: Example of a small part of a domain model (in
this case: HIV drug resistance) shown in a semantic inte-
gration notation. Specialization (hollow arrow) and com-
position (hollow rhombus) relations are applied, which is
typical of object-oriented modeling.
ViroLab project (Sloot et al., 2006). It deals with mu-
tations of HIV in human hosts and how these muta-
tions impact patient treatment options. The two most
critical relationships, inclusion and specialization, are
supported in our solution, in addition to primitive
datatypes and object arrays. The field list has been
abridged for reasons of clarity.
3.2 Modeling of Platform Capabilities
The true novelty of our approach is the concept of an
integration model. In contrast to the domain model
(which represents concepts from a specific area of hu-
man knowledge), the integration model represents ev-
erything that is connected with the process of build-
ing and maintaining an application for this domain:
it should model all elements of the application as re-
quired by the persistence layer, communication proto-
cols, computing data structures and all aspects which
are orthogonal to, and not directly dependent on the
problem domain. In order to model integration as-
pects we apply a similar object-oriented approach, al-
though integration models are usually more dynamic
(i.e. more code is used to implement operations com-
pared to static, descriptive domain models). While el-
ements of the domain model are called ”concepts”, we
refer to elements of the integration model as ”facets”.
Figure 2 presents the integration model for the
example mentioned above. In the ViroLab Virtual
Laboratory there is need to store pieces of valuable
data computed in the course of experimentation and
to record the ”provenance” of such data by using a
specialized tool (Bali
´
s et al., 2008). Note both fea-
tures (recording results and dispatching provenance
events) are unrelated to the application domain (HIV
drug-impeding mutations) and instead stem from sys-
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
76
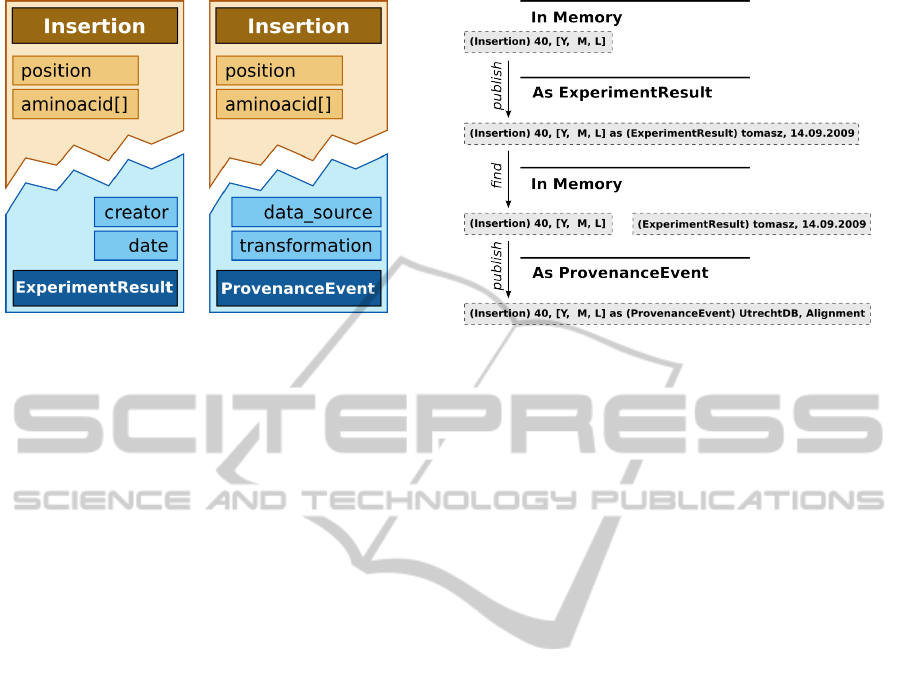
Figure 2: HIV drug resistance example (continued): a por-
tion of the integration model for this application. The ”In-
sertion” concept from the domain model is automatically
outfitted with the capability of being both an ”Experimen-
tResult” and a ”ProvenanceEvent” – the programmer may
use either one, according to the context of the given ”Inser-
tion” instance.
tem requirements associated with the virtual labora-
tory software.
There are two reasons behind constructing the in-
tegration model as an independent entity. First, the
separation serves the purpose of clarity, and enables
parallel evolution of both models, as explained in the
next paragraph. Maintenance also becomes easier (es-
pecially when the models become complex) and can
even be entrusted to separate developers, as the rea-
sons for extending either model are connected with
different sets of requirements.
The other reason is to stress two different evolu-
tion paths of these two fundamental elements. The
domain model grows together with the scope of the
knowledge body being incorporated in the system,
which is mainly caused by applying existing use cases
or features to new aspects of the scientists’ research
work. On the other hand, the integration model grows
along with new capabilities (new use cases or require-
ments) being implemented. Since each new feature
may be (and usually is) initially implemented for the
most stable parts of the domain model, the exten-
sion process for both models remains, to some de-
gree, independent. Of course, both paths can never be
separated completely, yet we argue that the proposed
method of separation delivers the much sought-after
flexibility of the constructed application.
3.3 ”Cartesian” Product of Facets
and Concepts for Integration
The final prerequisite of practical application of our
solution is a suitable mechanism for joining both
Figure 3: A single ”Insertion” instance might be used in dif-
ferent contexts during the lifespan of the application. Here,
two different elements of the environment use publishing
and querying ”ExperimentResults” for integration based on
the common understanding of the meaning of ”Insertion”.
The second element may subsequently publish such results
in another context (as ”ProvenanceEvents”).
models. The semantic integration framework per-
forms the juxtaposition dynamically and automati-
cally, with no need for programmer intervention. As
presented in Figure 2, as soon as both models are de-
ployed (presumably as parts of a larger application),
the developer links it with a provided library (where
the implementation of the semantic integration for a
given language resides) which mixes them both, pro-
ducing what may be referred to as a ”Cartesian prod-
uct model”. Every ”concept” of the domain model
may assume any ”facet” of the integration model –
both models are meant to be orthogonal.
The usual programming experience with models
involves handling objects in memory. Here, the pro-
grammer uses the domain model to represent aspects
of knowledge in terms of objects, creating hierarchies,
declaring fields etc. As soon as the concepts have to
act in a specific area of the application, the developer
uses one of their facets to perform the requested op-
erations. Examples include storing data in a database,
sending messages to another part of the application
(or to an external entity), presenting the user with a
dedicated UI and similar. As shown in Figure 3, the
concept object instantly acquires all the fields and op-
erations of a specific facet, so that it can be stored,
sent or presented accordingly, along with all facet-
related information. We call this action ”publishing
with a facet”.
If the nature of a facet allows for subsequent re-
trieval of the underlying concept, it can be located via
its associated facet data (see Figure 3). The frame-
work performs a decoupling action so that the user
receives the concept object and the facet object sepa-
SemanticIntegrationforModel-basedLifeScienceApplications
77

rately. Now (for instance) the user may apply the con-
cept object to perform publication with a different as-
pect of the integration model. This mechanism proves
very valuable for middleware elements of applications
which tend to work in multiple aspects, such as per-
sisting data in a DB or file, relaying it as events, pub-
lishing with a remote API or presenting to the user.
Technically, the generation of the dynamic Carte-
sian product and decoupling are effected using the
dynamic self-inspection capabilities of the Ruby pro-
gramming language
4
. Similar techniques might be
applied in any other modern programming platform
providing introspection. In fact, scripting languages
such as Python or Ruby are frequent programming
tools of choice in modern life sciences applications.
4 CASE STUDY – THE
POCKETFINDER
APPLICATION
This study focuses on applying the proposed seman-
tic integration methodology and tools to the domain
of bioinformatic protein research. This section ex-
plains the idea behind the PocketFinder application
and provides a detailed view on how the mechanisms
described in the previous sections were used to deliver
a final tool. In short, the main objective was to de-
velop a dedicated, efficient data store, populate it with
a large amount of computation results and present the
outcome in an interactive way to scientists. The re-
sulting application is called PocketFinder and is in-
tended for internal use by the bioinformatics group
at the Jagiellonian University Collegium Medicum in
Krakow.
4.1 Identification of Functional Sites
in Proteins
Understanding of how biological systems function is
the salient motivation for research in the field of bio-
chemistry and molecular biology. Proteins are the key
performers responsible for function at the molecular
level. Revealing their functional sites, i.e. parts of
their structure directly involved in performing their
function, is an important objective. The term “func-
tional site” is commonly used in reference to binding
sites or catalytic residues. Accordingly, experimental
and computational methods for identifying and char-
acterizing functional sites of proteins are being inten-
4
we used Ruby for our implementation of the semantic
integration idea
0 20 40 60 80 100 120 140 160 180
Residue number
-0.008
-0.006
-0.004
-0.002
0
0.002
0.004
0.006
0.008
0.01
∆H
~
-0.008
-0.006
-0.004
-0.002
0
0.002
0.004
0.006
0.008
0.01
Figure 4: DNA-3-methyladenine glycosylase I. (top) Rib-
bon structure with semitransparent spheres representing
atoms colored by ∆
˜
H values according to the scale pre-
sented in the profile below. (Bottom) Hydrophobicity dif-
ference (∆
˜
H) profile.
sively developed (Vajda and Guarnieri, 2006; Weigelt
et al., 2008).
Here we present a procedure developed to opti-
mize FOD (Fuzzy Oil Drop model) – an in silico
method applicable to prediction of functional sites in
proteins (Brylinski et al., 2007). The prediction relies
on hydrophobicity difference values (∆
˜
H) calculated
for each residue in a structure, based on FOD model
assumptions. The results for a given protein structure
are in the form of ∆
˜
H profile (Figure 4-top). The dis-
tribution of ∆
˜
H values within a structure can be rep-
resented as well (Figure 4-bottom).
The main goal was to establish optimal parame-
ters of the FOD method for prediction of catalytic
residues. The calculations were performed on a test
set comprising 521 enzyme chains. The obtained ∆
˜
H
values were treated as classifiers, discriminating cat-
alytic and non-catalytic residues. The actual class was
taken from CSA (Porter et al., 2004). As a result,
ROC curves were produced for each tested parameter
(in this case – hydrophobicity; Figure 5). Addition-
ally, areas bounded by the ROC curve (AUC) were
calculated.
4.2 Modeling Proteins and Profiles
We have applied the presented semantic integration
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
78

0
0.2
0.4
0.6
0.8
1
0 0.2 0.4 0.6 0.8 1
TPR
FPR
Aboderin
Brylinski
Figure 5: ROC curves created by thresholding the results
obtained for 24 hydrophobicity scales. Two curves refer-
ring to the best and original hydrophobicity scales are high-
lighted.
approach to developing domain and integration mod-
els for the storage of protein hydrophobicity profiles.
Out of necessity, modeling such entities proceeded
according to an iterative, sequential process, as user
requirements were analyzed and reflected in the data
model. It should be noted that the solution presented
in the above sections is readily applicable to such iter-
ative development, as the application developer may
freely alter the domain model without affecting the
underlying integration model. Moreover, any change
in the domain model may be easily propagated to the
user interface layer by modifying a dedicated REST-
ful user interface representing the application domain
(in this case – protein functional site study). Thus,
the overall framework may be adapted to the require-
ments of individual application domains while still re-
taining overall genericity (as reflected by the integra-
tion model).
Within the specific scope of the PocketFinder ap-
plication, each protein is decomposed into multiple
aminoacids, each of which may be associated with an
arbitrary number of measurements. Moreover, each
protein may correspond to a specific hydrophobicity
value and binding profile calculated on the basis of
multiple model types. This requirement gives rise to
a simple tree-like structure, which is depicted in Fig-
ure 6.
For PocketFinder we needed to prepare a data
store and ensure an efficient method of storing the
results of multiple computations running in parallel.
Two requirements needed to be fulfilled at this stage:
the repository had to fit in the available storage (thus,
the model had to store data in an efficient manner)
and the resulting tool had to ensure rapid access to
the stored data.
Figure 6: The FOD domain model (the most important part)
represents the central concept of protein profiles.
While forgoing traditional ACID guarantees, the
semantic integration tool nevertheless enables devel-
opers to implement OOP-like data models and store
them in an efficient manner. The final data repos-
itory for the Protein Pockets case study application
contains approximately 1.5 GB of data, yet still en-
ables rapid browsing with the use of the research and
validation tools presented below. Certain require-
ments introduced at the implementation stage were
translated into additional integration model features –
of note is the publish as uniqueitem operation which
implements the standard upsert behavior present in
many data storage systems. Thus, the semantic in-
tegration tool admits domain-mandated extensions to
its core functionality without the need to re-engineer
or refactor the system as a whole.
4.3 Creation of PocketFinder Research
and Validation Tool
Having developed and deployed the domain data
model and user interfaces, we proceeded with the
implementation of a research tool which would per-
form the scientific calculations involved in the Protein
Pockets case study. The application assumed the form
of a series of scripts, all of which cooperate to provide
the required functionality:
• A selection of Ruby modules to populate the ini-
tial data store with information on proteins and
their constituent aminoacids;
• A Python experiment used to generate and execute
a sequence of scripts in a parameter-study mode,
where each of the launched scripts operates sep-
arately while retrieving data from the semantics
store;
• Perl scripts which implement the specific func-
tionality of the PocketFinder application, i.e.
SemanticIntegrationforModel-basedLifeScienceApplications
79

calculate measurement values for individual
aminoacids according to predefined models, and
commit the results to the data store.
The RESTful domain interface (which is
application-specific and should be considered part
of the application itself), makes it easy to interface
the semantic integration tool from any programming
language equipped with a HTTP communications
library (which includes a vast majority of modern
languages). If we count shell scripting, the Pock-
etFinder application itself comprises four different
programming languages, all of which communicate
using the semantic integration tools.
5 OTHER APPLICATION AREAS
The presented solution was applied to several other
use cases, one of which was the HIV mutation and
drug resistance problem (Gubala et al., 2009) – in
fact, the requirements of that project were the driv-
ing force behind the semantic integration idea pre-
sented in this paper. Apart from the PocketFinder
application, we have also applied refined method-
ologies in the computational chemistry domain (as
a semantic metadata engine in the InSilicoLab por-
tal (Kocot et al., 2012)
5
) and crisis warning and
management systems (as a semantic integration layer
for metadata and registries of dike sensors moni-
tored by the UrbanFlood EWS system (Balis et al.,
2011)
6
). Currently, we continouosly refine and apply
the methodology to further science-related domains:
multiphisics and multiscale simulations in the scope
of MAPPER project (Rycerz and Bubak, 2011)
7
and
complex applications related to the Virtual Physio-
logical Human domain in the scope of VPH-Share
project (Nowakowski et al., 2012)
8
.
The development of these systems highlighted
good reusability of integration models (see 3.2). Once
defined, facets such as ”UniqueInput”, ”Versioned-
Document” and ”JSONObject” tend to be applicable
to a variety of different realizations – this is due to
the relatively similar needs of users from different do-
mains, especially regarding the rather low level of the
software stack (persistence, presentation, communi-
cation). Here, programmers have greater freedom to
5
InSilicoLab e-science application portal, http://
insilicolab.cyfronet.pl/
6
Common Information Space, http://urbanflood.
cyfronet.pl/
7
Multiscale Applications on European e-Infrastructures,
http://www.mapper-project.eu/
8
VPH-Share Sharing for Healthcare - A Research Envi-
ronment, http://www.vph-share.eu/
adopt preferred solutions, so we opted for reusabil-
ity wherever the application requirements did not pre-
clude it. We believe that this observation validates our
core decision to separate the domain and integration
models.
6 SUMMARY
Summarizing, the ability to implement novel,
mission-critical tools in three different domains (in-
cluding HPC-based computational science and real-
time monitoring) in a six-month time frame by a team
of three programmers (not including end users who
contributed steady feedback) can be taken as evidence
for the extensive potential of the proposed method-
ology and accompanying technologies. It should be
noted, however, that when it comes to programmer
productivity or the agility of the software implemen-
tation process, it is difficult to provide any convinc-
ing numerical measurements or metrics – particularly
ones that would involve semantics and their impact
on the process. Results need to be judged on the ba-
sis of common sense and past experience – yet the
outcome of our to-date research and development ef-
forts is a clear sign that the semantic integration idea
covered in this article is a worthwhile proposition for
e-science researchers and scientific developers alike.
ACKNOWLEDGEMENTS
The research presented in this paper has been par-
tially supported by the European Union within the
EU Project MAPPER (grant no. RI-261507) and the
EU Project VPH-Share (grant no. 269978). It was
also supported by the Polish national PL-Grid Project
POIG.02.03.00-00-007/08-00 and KI IET AGH grant.
REFERENCES
Bali
´
s, B., Bubak, M., Pelczar, M., and Wach, J. (2008).
Provenance tracking and querying in the virolab vir-
tual laboratory. In Proceedings of the 8th IEEE In-
ternational Symposium on Cluster Computing and the
Grid 2008 (CCGRID), pages 675–680. IEEE Com-
puter Society.
Balis, B., Kasztelnik, M., Bubak, M., Bartynski, T., Gubala,
T., Nowakowski, P., and Broekhuijsen, J. (2011). The
urbanflood common information space for early warn-
ing systems. In Proceedings of the International Con-
ference on Computational Science ICCS 2011, vol-
ume 4, pages 96–105. Procedia Elsevier.
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
80

Brylinski, M., Prymula, K., Jurkowski, W., Kochanczyk,
M., Stawowczyk, E., Konieczny, L., and Roterman,
I. (2007). Prediction of functional sites based on the
fuzzy oil drop model. PLoS Comput Biol, 3(5):e94.
Bubak, M., Szepieniec, T., and Wiatr, K., editors (2012).
Building a National Distributed e-Infrastructure -
PL-Grid - Scientific and Technical Achievements.
Springer.
Bubak, M. and Unger, S., editors (2007). KWf-Grid -
The Knowledge-based Workflow System for Grid Ap-
plications. Academic Computer Centre CYFRONET
AGH.
Cannataro, M., Guzzi, P., Mazza, T., Tradigo, G., and Vel-
tri, P. (2007). Using ontologies for preprocessing and
mining spectra data on the grid. Future Generation
Computer Systems, 23(1):55–60.
Corcho, O., Alper, P., Kotsiopoulos, I., Missier, P., Bech-
hofer, S., and Goble, C. (2006). An overview of s-
ogsa: A reference semantic grid architecture. Web Se-
mantics: Science, Services and Agents on the World
Wide Web, 4(2):102–115.
Gubala, T., Bubak, M., and Sloot, P. M. (2009). Semantic
Integration of Collaborative Research Environments,
chapter XXVI, pages 514–530. Information Science
Reference IGI Global.
Gubala, T. and Hoheisel, A. (2006). Highly dynamic work-
flow orchestration for scientific applications. In Core-
GRID Intergation Workshop 2006 (CIW06), pages
309–320. ACC CYFRONET AGH.
Guru, S. M., Kearney, M., Fitch, P., and Peters, C. (2009).
Challenges in using scientific workflow tools in the
hydrology domain. In 18th World IMACS and MOD-
SIM09 Int. Congress on Modelling and Simulation,
pages 3514–3520. Modelling and Simulation Society
of Australia and New Zealand.
Kocot, J., Szepieniec, T., Harezlak, D., Noga, K., and
Sterzel, M. (2012). InSilicoLab Managing Com-
plexity of Chemistry Computations, pages 265–275.
Springer.
Kondylakis, H., Analyti, A., and Plexousakis, D. (2007).
Quete: Ontology-based query system for distributed
sources. Lecture Notes in Computer Science,
4690:359–375.
Kryza, B., Slota, R., Majewska, M., Pieczykolan, J., and Ki-
towski, J. (2007). Grid organizational memory – pro-
vision of a high-level grid abstraction layer supported
by ontology alignment. Future Generation Computer
Systems, 23(3):348–358.
Nowakowski, P., Bartyski, T., Gubaa, T., Harlak, D.,
Kasztelnik, M., Meizner, J., and Bubak, M. (2012).
Managing cloud resources for medical applications.
In Proceedings of Cracow Grid Workshop 2012
(CGW’12), pages 35–36. ACC Cyfronet AGH.
Petrie, C. (2009). The semantics of ”semantics”. IEEE
Internet Computing, 13(5):96–95.
P.M. Gordon, Q. Trinh, C. S. (2007). Semantic Web Ser-
vice provision: a realistic framework for Bioinformat-
ics programmers. Bioinformatics, 23(9):1178–1180.
Porter, C. T., Bartlett, G. J., and Thornton, J. M. (2004). The
Catalytic Site Atlas: a resource of catalytic sites and
residues identified in enzymes using structural data.
Nucleic Acids Res, 32(Database issue):D129–D133.
Rodriguez, M. and Egenhofer, M. (2003). Determiniing
semantic similarity among entity classes from differ-
ent ontologies. IEEE Transactions on Knowledge and
Data Engineering, 15(2):442–456.
Rycerz, K. and Bubak, M. (2011). Building and Run-
ning Collaborative Distributed Multiscale Applica-
tions, chapter 6, pages 111–130. J. Wiley and Sons.
Sloot, P. M. A., Tirado-Ramos, A., Altintas, I., Bubak, M.,
and Boucher, C. (2006). From molecule to man: De-
cision support in individualized e-health. Computer,
39(11):40–46.
Vajda, S. and Guarnieri, F. (2006). Characterization of
protein-ligand interaction sites using experimental and
computational methods. Curr Opin Drug Discov De-
vel, 9(3):354–362.
Weigelt, J., McBroom-Cerajewski, L. D. B., Schapira, M.,
Zhao, Y., and Arrowmsmith, C. H. (2008). Structural
genomics and drug discovery: all in the family. Curr
Opin Chem Biol, 12(1):32–39.
SemanticIntegrationforModel-basedLifeScienceApplications
81
