
Chemoinformatics in Drug Design. Artificial Neural Networks for
Simultaneous Prediction of Anti-enterococci Activities and
Toxicological Profiles
Alejandro Speck-Planche and M. N. D. S. Cordeiro
REQUIMTE/Department of Chemistry and Biochemistry, University of Porto, 4169-007 Porto, Portugal
Keywords: Artificial Neural Networks, Enterococci, Inhibitors, Toxicity, Topological Indices, mtk-QSBER, BC-3781.
Abstract: Enterococci are dangerous opportunistic pathogens which are responsible of a huge number of nosocomial
infections, displaying intrinsic resistance to many antibiotics. The battle against enterococci by using
antimicrobial chemotherapies will depend on the design of new antibacterial agents with high activity and
low toxicity. Multi-target methodologies focused on quantitative-structure activity relationships (mt-
QSAR), have contributed to rationalize the process of drug discovery, improving the knowledge about the
molecular patterns related with antimicrobial activity. Until know, almost all mt-QSAR models have
considered the study of biological activity or toxicity separately. Here, we developed a unified mtk-QSBER
(multitasking quantitative-structure biological effect relationships) model for simultaneous prediction of
anti-enterococci activity and toxicity on laboratory animal and human immune cells. The mtk-QSBER
model was created by using artificial neural network (ANN) analysis combined with topological indices,
with the aim of classifying compounds as positive (high biological activity and/or low toxicity) or negative
(otherwise) under multiple experimental conditions. The mtk-QSBER model correctly classified more than
90% of the whole dataset (more than 10900 cases). We used the model to predict multiple biological effects
of the investigational drug BC-3781. Results demonstrate that our mtk-QSBER may represent a new
horizon for the discovery of desirable anti-enterococci drugs.
1 INTRODUCTION
The genus Enterococcus is formed by a group of
low-GC Gram-positive, catalase-negative, non-
spore-forming, facultative anaerobic bacteria that
can occur both, as single cocci and in chains (Fisher
and Phillips, 2009). Several species belonging to
Enterococcus spp. are opportunistic pathogens
which constitute the major cause of nosocomial
infections such as bacteremia, bacterial endocarditis,
diverticulitis, meningitis and urinary tract infections
(Ryan and Ray, 2004). The successful elimination of
infections produced by enterococci will depend on
two very important aspects: the efficiency of the
antimicrobial chemotherapies used against the
infection and the safety of the drugs for human
health.
Antimicrobial chemotherapies against
Enterococcus spp. are focused on the use of the β-
lactam antibiotic ampicillin or combination of a cell
wall–active agent (such as ampicillin or
vancomycin) with aminoglycosides (gentamicin,
tobramycin), which may result in synergistic
bactericidal activity against enterococci (Ryan and
Ray, 2004). However, enterococci are intrinsically
resistant to a broad range of antibiotics commonly
used in the hospital setting, which explains in some
way, the high prevalence of these bacteria in
nosocomial infections (Brachman and Abrutyn,
2009). The most alarming aspect in enterococci is
that they are reservoirs for antibiotic resistance
genes, as may be exemplified by their ability to
transfer vancomycin resistance to methicillin-
resistant Staphylococcus aureus (MRSA), for which
vancomycin remains the last therapeutic alternative
(Figure 1). For this reason, there is an increasing
necessity for the search of new, potent and more
efficient antibacterial chemotherapies against
enterococci. On the other hand, when any
antibacterial drug is designed, serious concerns are
expected due to its appearance of toxic effects on
human health. Thus, many trials are carried out on
laboratory animals.
In this sense, Mus musculus and Rattus
458
Speck-Planche A. and Cordeiro M..
Chemoinformatics in Drug Design. Artificial Neural Networks for Simultaneous Prediction of Anti-enterococci Activities and Toxicological Profiles.
DOI: 10.5220/0004542004580465
In Proceedings of the 5th International Joint Conference on Computational Intelligence (NCTA-2013), pages 458-465
ISBN: 978-989-8565-77-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
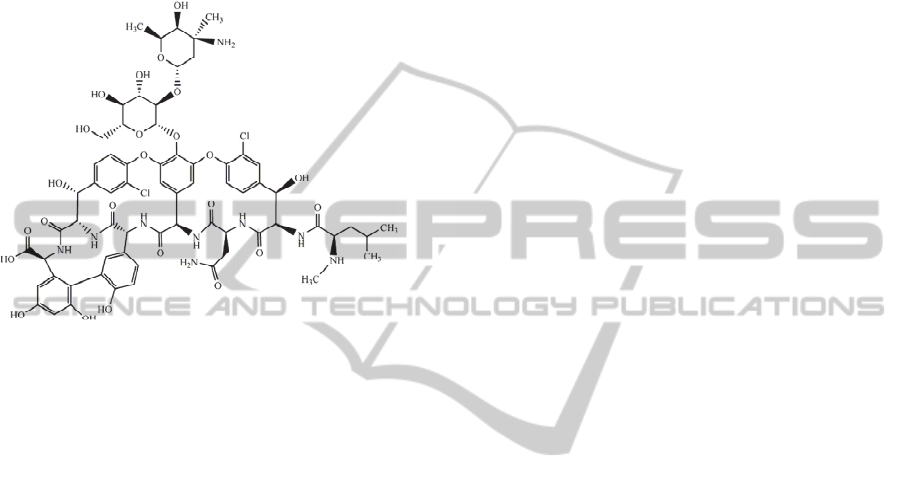
norvegicus are the most valuable species (Hau and
Schapiro, 2011), suffering as consequence of endless
batteries of toxicity tests. At the same time, the study
of the effects of chemicals on human immune
system cells is also very important because these are
the lines of defense of the human body, protecting it
against the entry of any foreign agent (Flaherty,
2012).
Figure 1: Vancomycin: one of the most powerful broad
spectrum antibacterial drugs.
In the last six years, several researchers have
emphasized the use of multi-target for quantitative-
structure activity relationships (mt-QSAR), which
have emerged as very useful tools for rational design
and virtual screening of compounds with dissimilar
biological activities, by considering many different
biological targets (biomolecules, cell lines, tissues,
organisms) (Munteanu et al., 2009; Prado-Prado et
al., 2009; Speck-Planche and Kleandrova, 2012;
Speck-Planche et al., 2012), to the assessment of
pharmacological/toxicological profiles in multiple
assay conditions (Speck-Planche et al., 2013;
Tenorio-Borroto et al., 2012).
Nowadays, no methodology has been reported
for the prediction of anti-enterococci activity and
toxicity at the same time. Furthermore, sometimes,
non-linear modeling by using pattern recognition
methods such as Artificial Neural Networks (ANN)
(Prado-Prado et al., 2010; Tenorio-Borroto et al.,
2012), should be considered in order to find better
relationships between the molecular descriptors
describing the chemical structure of the compounds,
and their biological activities and/or toxicities. Thus,
with the objective to reduce the high costs of
experimentation, in this work we introduce the first
unified multitasking model based on quantitative-
structure biological effect relationships (mtk-
QSBER) and ANN analysis, for the simultaneous
prediction of anti-enterococci activities and
toxicological profiles in multiple assay conditions.
2 MATERIALS AND METHODS
2.1 Topological Indices
Molecular descriptors have served as essential
support for the development and consolidation of
important disciplines such as chemoinformatics
(Oprea, 2005). Among them, topological indices
(TIs) have been very useful to correlate the chemical
structure of compounds with the pharmacological
activity (QSAR) or with the toxicity (QSTR)
(Todeschini and Consonni, 2009). Descriptors like
TIs can be considered as numerical parameters of a
graph which characterize its topology, being graph
invariants, i.e., they will never depend on how the
graph (molecule) will be drawn and/or enumerated
(Todeschini and Consonni, 2009). Then, the
topology of a molecule can be studied in terms of its
size (volume), molecular accessibility, shape,
electronic factors and many other properties. For
development of this work, we selected some of the
classical TIs which include valence connectivity
indices (Kier and Hall, 1986), bond connectivity
indices (Estrada, 1995), and Balaban index
(Balaban, 1982).
2.2 Dataset: Calculation of the
Descriptors and Development of the
mtk-QSBER Model
One of the main factors to take into account for the
development of a predictive model is the use of an
appropriate dataset. In this sense, we extracted a raw
dataset from the large and highly accurate database
CHEMBL (Gaulton et al., 2012), which is available
at http://www.ebi.ac.uk/chembldb/. We retrieved
13073 endpoints of different biological effects
reported for more than 9000 compounds. With the
aim of reducing the uncertainty of the data, we
deleted all the endpoints with missing values or units
of biological effects. After that, our dataset was
formed by N
c
= 8560 compounds, being some of
them tested against more than one experimental
condition c
j
. For this reason, the dataset contained
10918 statistical cases. To develop the mtk-QSBER
model, we employed a similar methodology to that
reported for the for the simultaneous modeling of
antituberculosis activity and toxic effects on
laboratory animals (Speck-Planche et al., 2013). As
stated in these previous works, any set of
ChemoinformaticsinDrugDesign.ArtificialNeuralNetworksforSimultaneousPredictionofAnti-enterococciActivities
andToxicologicalProfiles
459

experimental conditions c
j
by which a compound is
tested, can be expressed as an ontology c
j
=> (m
e
, b
t
,
a
i
, l
c
). In this ontology, m
e
represents the measure of
biological effect (anti-enterococci activities or
toxicity). The element b
t
is referred to different
biological targets such as enterococci, Mus musculus
and Rattus norvegicus, and human immune system
cells (lymphocytes). For all biological targets,
information about different strains was taken into
consideration. The element a
i
defines specific
information regarding a test, i.e., if an assay is
focused on the study of functional (F) or
pharmacokinectic/pharmacodynamic profiles (A).
The term l
c
is the level of curation or verification of
the experimental information provided by a
particular test. The elements m
e
, b
t
, a
i
, and l
c
define
the four conditions which can change in our dataset.
So, we had N = 10918 cases from N
c
= 8560
compounds mentioned above, where the
experiments were performed using at least one out
of Nm
e
= 18 measures of biological effects, against
at least one out of Nb
t
= 131 biological targets, in
one out of Na
i
= 2 different types of assay
information, with at least one out of Nl
c
= 3 levels of
curation of the experimental information. In the case
of the element m
e
, we had diverse measures of
biological effects which were expressed in different
units. For this reason, all values of antibacterial
activity against enterococci were converted to nmol/l
(nM), while all toxicity values associated with
laboratory animals were expressed in umol/kg
(micromoles per kilograms). In both kinds of
conversions, it was necessary to divide the value of
each compound by its molar mass, and after multiply
by a factor (usually 10
3
or 10
6
). We realized these
transformations in order to make a better
interpretation of the biological data which permitted
us a more rigorous comparison between the
biological effects of any two compounds, measured
under exactly the same set of conditions c
j
. Data
associated with cytotoxicity against immune cells,
remained in nM. These transformations together
with the element l
c
, also contribute significantly to
reduce and control data uncertainty. All cases in our
dataset were assigned to 1 out of 2 possible groups
related with the biological effect of a defined
compound i in a specific condition c
j
[BE
i
(c
j
)]. Then,
any compound was considered as positive [BE
i
(c
j
) =
1] when it had high anti-enterococci activity, or any
desirable toxicological profile, otherwise, the
compound was considered as negative [BE
i
(c
j
) =
1]. All assignments were realized taking into
account certain cutoff values of biological effects
which are depicted in Table 1. For the whole dataset,
we used a file containing the SMILES of the
compounds/cases. Calculation of TIs using SMILES
was performed with the software MODESLAB
version 1.5 (Estrada and Gutiérrez, 2002-2004). Our
intention is to predict the biological effect of any
compound depending on the molecular structure and
the experimental conditions c
j
. For this reason if we
use the original TIs calculated above, they will not
discriminate the biological effect for a given
compound by varying the different conditions c
j
. To
achieve that goal, and inspired by the use of the
moving average approach (MAA) (Hill and Lewicki,
2006), we introduced new sets of molecular
descriptors like TIs which can be defined according
to the following equation:
ΔTI
i
(c
j
) = TI
i
– avgTI
i
(c
j
) (1)
In Eq. 1, the descriptor avgTI
i
(c
j
) characterizes each
set G of compounds tested under the same
experimental condition c
j
, being calculated as the
sum of all the TI
i
values for compounds in a subset
of G, which were considered as positive cases
[BE
i
(c
j
) = 1] in the same element of the ontology
(experimental condition) c
j
. For example, in the case
of the element b
t
, the descriptor avgTI
i
(c
j
) for a set G
of compounds tested against a defined target b
t
(bacterial strain, immune cell, etc), was calculated as
the average of the TI
i
by considering only the subset
of G, i.e., those compounds which were considered
as positive [BE
i
(c
j
) = 1]. A similar procedure was
carried out for the elements m
e
, a
i
, and l
c
. Anyway,
in Eq. 1, the most important element is the
descriptor ΔTI
i
(c
j
), which considers both, the
molecular structure and the experimental conditions
c
j
. For this reason, descriptors of the form ΔTI
i
(c
j
)
(120 in total) were used to develop the mtk-QSBER
model. These descriptors represent the deviation (in
structural terms) of a compound from the positive
compounds. The CHEMBL codes, SMILES and
other relevant experimental data for all the
compounds used in this work, appear in the
Supplementary Information 1 (Suppl. Inf. 1) file.
Our dataset of 10918 cases was randomly split into
two series: training and prediction sets. The training
set was used to construct the mtk-QSBER model.
This was formed by 8298 cases, with 4217 of
them considered as positive and 4081 negative. The
prediction set was used for validation of the model
and assessment of its predictive power, being
composed by 2620 cases, 1353 positive and 1267
negative cases. Taking into consideration that large
number of molecular descriptors, we used a
combination of the attribute evaluator
CFsSubsetEval and the search algorithms called
IJCCI2013-InternationalJointConferenceonComputationalIntelligence
460

Table 1: Cutoff values for diverse measures of biological effects.
Standard measure
(units)
a
Biological profile Description Cutoff
b
CC
50
(nM) Cytotoxicity Concentration required to reduce cell viability by 50% ≥70000
ED
50
(umol/kg)
In vivo antibacterial
activity
Concentration required to produce a specific effect in
half of an animal population comprising a test sample
≤14.53
IC
50
(nM) Antibacterial activity
Concentration required to inhibit the growth of a
microorganism by 50%
≤836.21
LD
50
(umol/kg)im Toxicity Lethal dose at 50% after intramuscular administration ≥960
LD
50
(umol/kg)ip Toxicity Lethal dose at 50% after intraperitoneal administration ≥1050
LD
50
(umol/kg)iv Toxicity Lethal dose at 50% after intravenous administration ≥502.12
LD
50
(umol/kg)oral Toxicity Lethal dose at 50% after oral administration ≥1110
LD
50
(umol/kg)sc Toxicity Lethal dose at 50% after subcutaneous administration ≥713.58
MBC (nM) Antibacterial activity Concentration required to kill 100% of microorganisms ≤11040.2
MIC (nM) Antibacterial activity
Lowest concentration that prevents the visible growth of
a microorganism
≤6000
MIC
50
(nM) Antibacterial activity
Minimum inhibitory concentration required to inhibit
the growth of 50% of microorganisms
≤2112.21
MIC
90
(nM) Antibacterial activity
Minimum inhibitory concentration required to inhibit
the growth of 90% of microorganisms
≤4982.18
ND
50
(umol/kg)ip Toxicity
Dose causing a neurological deficit in 50% of
organisms after intraperitoneal administration
≥239.71
ND
50
(umol/kg)oral Toxicity
Dose causing a neurological deficit in 50% of
organisms after oral administration
≥375.52
PI
Activity/Toxicity Protective index ≥4.9
TD
50
(umol/kg)ip Toxicity
Dose at which toxicity occurs in 50% of organisms after
intraperitoneal administration
≥395.21
TD
50
(umol/kg)oral Toxicity
Dose at which toxicity occurs in 50% of organisms after
oral administration
≥632.5
TD
50
(umol/kg)sc Toxicity
Dose at which toxicity occurs in 50% of organisms after
subcutaneous administration
≥1144.41
a
Referred to the element m
e
of the ontology c
j
.
b
Necessary condition for considering a compound as positive.
BestFirst and GeneticSearch, all of them
implemented in the program WEKA version 3.6.9
(Hall et al., 1999-2013). The purpose was to reduce
the dimensionality, i.e., the number of molecular
descriptors. We took into account that at least one
descriptor representing each element of the ontology
c
j
, must be selected. To seek the best mtk-QSBER
model, ANN analysis was performed using the
software STATISTICA 6.0 (StatSoft, 2001). In order
to select the most important descriptors, a sensitivity
analysis was performed. In this sense, the neural
network module of STATISTICA has defined a
missing value substitution procedure, which is used
to allow predictions to be made in the absence of
values for one or more input variables (StatSoft,
2001). Thus, to define the sensitivity of a particular
input variable (descriptor) v, each ANN is run on a
defined set of cases (training cases), where a
network error is accumulated (Hill and Lewicki,
2006). After, the network is run again using the
same cases, but this time replacing the observed
values of v with the value estimated by the missing
value procedure. So, a new network error is
accumulated. Taking into consideration that some
information that each network uses, has effectively
been removed (i.e. one of the input variables), it is
logical to expect some deterioration in error to occur
(Hill and Lewicki, 2006). Then, the sensitivity of
any input variable is calculated as the ratio of the
error with missing value substitution to the original
error. The more sensitive the network is to a
particular input variable (descriptor), the greater the
deterioration we can expect, and therefore the
greater the ratio. This procedure used to detect the
relative importance of a variable, is efficiently
implemented in STATISTICA 6.0 (StatSoft, 2001).
We need to emphasize that only the variables with
high sensitivity values (>1) were selected, and we
ensured that at least one variable belonging to each
element of the ontology c
j
was among the chosen
variables in the final model. The quality and
predictive power of our mtk-QSAR model by
examining some statistical indices such as the
sensitivity (Sens) and specificity (Spec), the
Mathew's correlation coefficient (MCC), and the
areas under the receiver operating characteristic
ChemoinformaticsinDrugDesign.ArtificialNeuralNetworksforSimultaneousPredictionofAnti-enterococciActivities
andToxicologicalProfiles
461

(ROC) curves (Speck-Planche et al., 2012) in both,
training and prediction sets. When the analyst does
not know the system a priori, very sophisticated
methods to seek the best descriptors and optimize of
the neural networks may be needed. However,
taking in mind that the dataset was rigorously
curated, and that descriptors of type ΔTI
i
(c
j
) can
phenomenologically explain the structural variation
in the dataset, simple rules for optimizing neural
network can be applied. Thus, the Intelligent
Problem Solver was used to seek the best networks.
This module provides the search for the best models,
and through an internal algorithm, it considers the
maximum number of hidden units, based on the
number of variables and cases (for each type of
network architecture) (StatSoft, 2001).
The first five runs served to determine the best
type of neural network. After, the number of hidden
units of the network selected as the best was
employed as maximum number of hidden units in
five new runs. The process was repeated until a
network had enough small number of hidden units
and the total percentage of correct classification
(accuracy) of cases was ≥90% in both training and
prediction sets.
3 RESULTS AND DISCUSSION
For the selection of the best mtk-QSBER model we
analyzed the different types of ANNs. These were
linear neural network (LNN), probabilistic neural
network (PNN), radial basis function (RBF), and
multilayer perceptron (MLP). We also took into
consideration the principle of parsimony, which
means that the model with the highest statistical
quality, but having as few variables as possible,
should be selected. As depicted in Table 2, the best
mtk-QSBER model found by us was that associated
with the RBF-ANN, which displays the highest
performance in terms of sensitivity and specificity,
with the lowest errors when compared with the other
ANNs. The profile of this ANN is: RBF 5:5-767-
1:1. The symbologies for all the descriptors together
with their corresponding meanings appear
represented in Table 3. Our mtk-QSBER model,
could correctly classify 7740 out of 8298 cases were
correctly classified, for an accuracy of 93.28% in
the training set, while in prediction set, 2395 out of
2620 cases were correctly classified and the
value of accuracy was 91.41%. More details about
the results of classification and predictions can be
found in Table 4 and Supplementary Information 2
(Suppl. Inf. 2) file respectively. All the average
descriptors used in this work, together with the
percentages of correct classification depending on
the elements m
e
, b
t
, a
i
, and l
c
, can be found in
Supplementary Information 3 (Suppl. Inf. 3) .
The values of areas under ROC curves played an
important role to confirm the quality and the
predictive power of the model. The values of area
under the ROC curve were 0.981 and 0.965 for
training and prediction sets respectively (Figure 2).
These values of area can be interpreted as follows:
value of area 0.981, means that a randomly selected
compound or case from the active group (protein
inhibitor) will have a larger value of probability than
a randomly selected compound or case from the
inactive group, 98.1% of the times. A similar
deduction can be made from the area under the ROC
curve for the case of the prediction set. We are
demonstrating that our mtk-QSBER model is not a
random classifier because the areas under the ROC
curves are clearly different from those obtained by
random classifiers (area = 0.5). By analyzing the
results of Table 4 and the values of areas under the
ROC curves, we can say that our mtk-QSBER model
has excellent quality and predictive power which is
comparable with other reports in the literature
related to the use of the mt-methodologies combined
with ANN analysis (Prado-Prado et al., 2010;
Tenorio-Borroto et al., 2012). The use of classical
TIs permits to obtain simple substructural and
physicochemical information. One important aspect
is that all descriptors employed to construct the mtk-
Table 2: Performance of the different ANNs.
CHARACTERISTICS
Symbology LNN MLP (TLP)
a
MLP (FLP)
b
RBF PNN
Profile
5:5-1:1 5:5-8-1:1 5:5-7-10-1:1
5:5-767-1:1
5:5-8298-2-2:1
Training set
Sens (%)
58.88 72.11 77.35
92.98
95.04
Spec (%)
58.61 72.38 77.82
93.58
27.96
Prediction set
Sens (%)
58.17 72.28 77.01
90.76
94.38
Spec (%)
61.01 73.72 78.06
92.11
28.02
a
Abbreviation for three layer perceptron ANN.
b
Nomenclature referred to four layer perceptron ANN.
IJCCI2013-InternationalJointConferenceonComputationalIntelligence
462
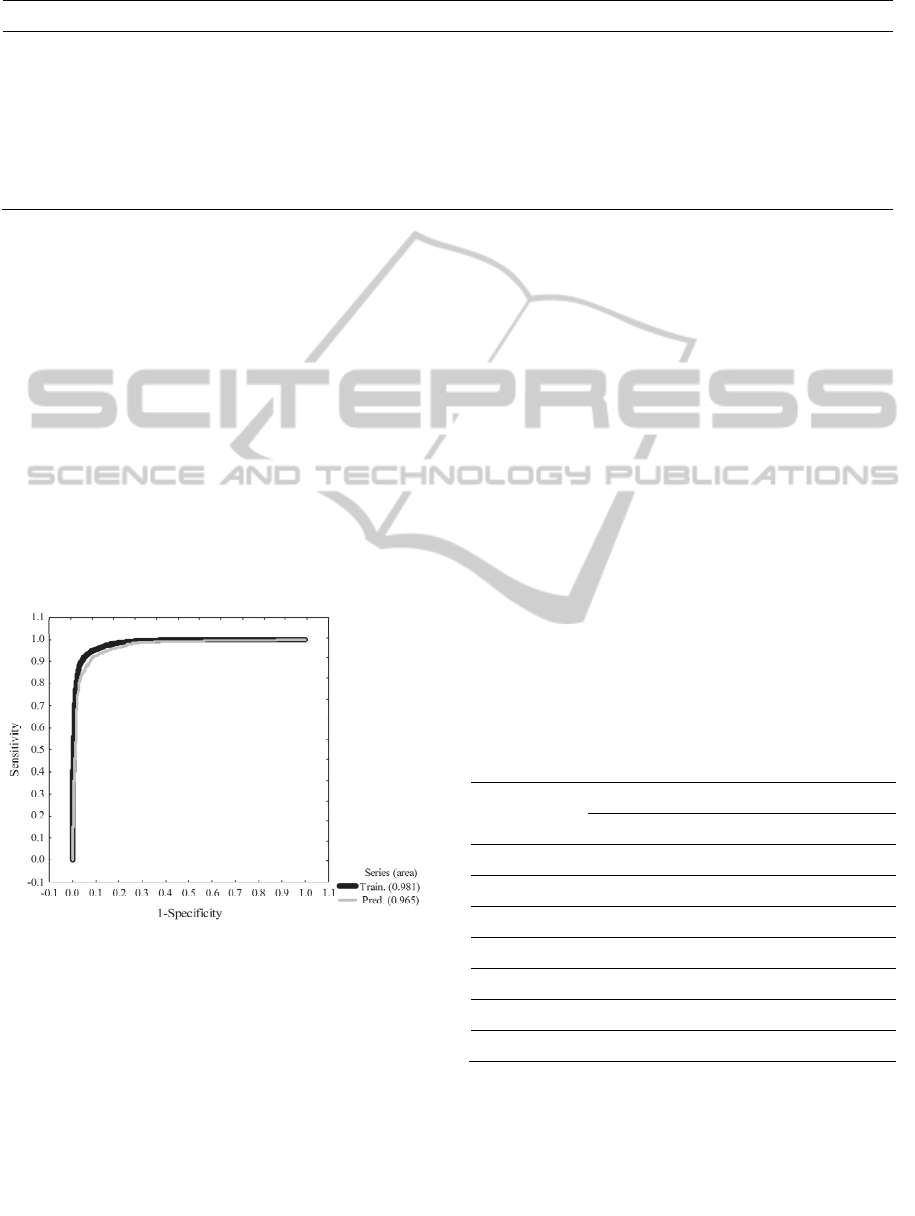
Table 3: Descriptors used to construct the mtk-QSBER model.
Descriptor Concept
Δ[
3
χ
v
p
(m
e
)]
Deviation of the vertex connectivity index of order 3 and type path, dependent on the molecular structure and the
measure of biological effect
Δ[
2
e
p
(b
t
)]
Deviation of the bond connectivity index of order 2 and type path, dependent on the molecular structure and the
biological target
Δ[
6
e
ch
(b
t
)]
Deviation of the bond connectivity index of order 6 and type chain (ring), dependent on the molecular structure and
the biological target
ΔJ(a
i
) Deviation of the Balaban index, dependent on the molecular structure and the assay information
Δ[
5
e
ch
(l
c
)]
Deviation of the bond connectivity index of order 5 and type chain (ring), dependent on the molecular structure and
the level of curation of the experimental information
QSBER model have the form Δ
TI
i
(c
j
). These
descriptors can be considered as measures of the
similarity/dissimilarity of a given compound respect
a group of positive cases depending on the molecular
structure, and a specific element of the ontology c
j
(m
e
, b
t
, a
i
, or l
c
). Thus, the descriptor Δ[
3
χ
v
p
(m
e
)]
encodes information related with the molecular
accessibility in those regions which contain linear
fragments formed by three bonds (Estrada, 2002).
This variable takes into consideration the structure of
the molecule and the measure of biological effect
which was used for that molecule. The variable
Δ[
2
e
p
(b
t
)], is strongly related with the molecular
volume in linear substructures containing two bonds
(Estrada, 1995).
Figure 2: ROC curves for the mtk-QSBER model.
A similar physicochemical information is
encoded by the descriptor Δ[
6
e
ch
(b
t
)], but with the
difference that only regions formed by six-
membered rings are taken into account. The variable
Δ[
2
e
p
(b
t
)] as well as Δ[
6
e
ch
(b
t
)] depend on the
chemical structure and the biological target against
which a compound was tested. The variable, ΔJ(a
i
)
is focused on the global shape (Balaban, 1982),
depending on the structure of the compound and the
assay information. Finally, Δ[
5
e
ch
(l
c
)] will depend on
the molecular structure (considering heteroatoms)
and the level or degree of curation of the
experimental information, and its structural
information will be concerned with the molecular
volume in those regions with five-membered rings.
Any model should be able to predict compounds
which were not used either training or prediction
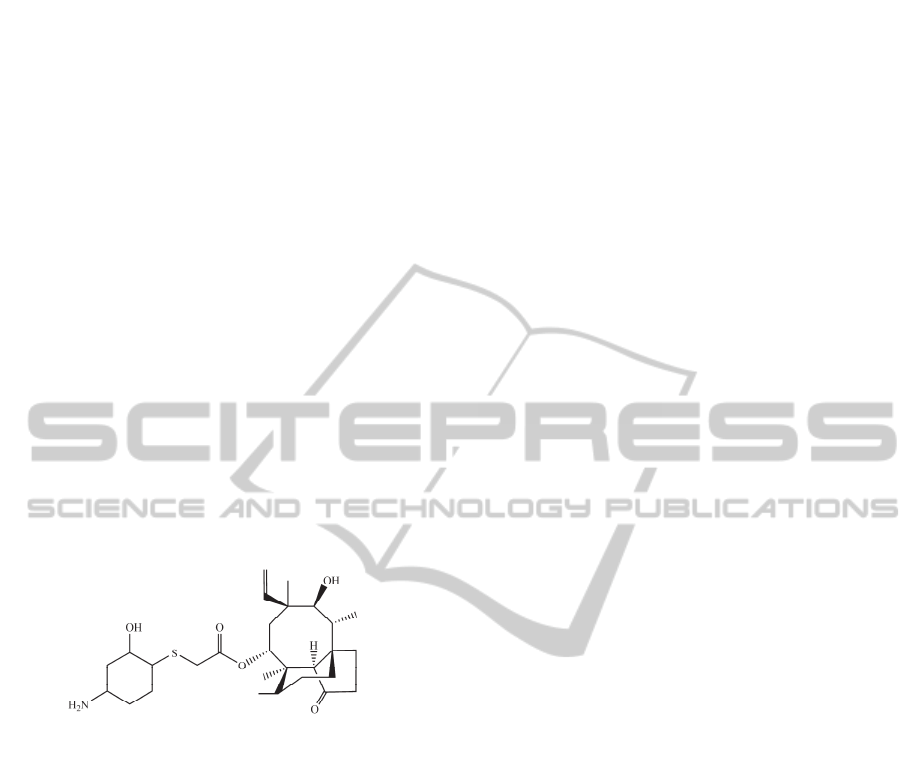
sets. For this reason, in order to show how our mtk-
QSBER model works, we predict the effects of the
antibiotic BC-3781 against enterococci, as well as
different toxicological profiles under diverse
experimental conditions. BC-3781 is an
investigational drug (Figure 3), which has being
studied as a broad spectrum antibacterial agent due
to its activity against Gram-positive cocci,
Haemophilus influenzae, and many other bacteria
which cause serious skin infections, bacterial
pneumonia or opportunistic infections. BC-3781 has
been obtained by Nabriva Therapeutics (Sader et al.,
2012), a company focused on developing new class
of antibiotics against serious bacterial infections.
Table 4: Results of classification.
Classification
Training set Prediction set
Positive Negative Positive Negative
Total
4217 4081 1353 1267
Correct
a
3921 3819 1228 1167
Wrong
296 262 125 100
Correct (%)
b
92.98 93.58 90.76 92.11
Wrong (%)
7.02 6.42 9.24 7.89
Acc (%)
c
93.28 91.41
MCC
0.866 0.828
a
Compounds which were correctly classified by the model.
b
Formally known as sensitivity (Sens) for positive cases and
specificity (Spec) for negative.
c
Referred to the accuracy as total percentage of correct
classification.
All information regarding this systemic product
can be found at http://www.nabriva.com/.
ChemoinformaticsinDrugDesign.ArtificialNeuralNetworksforSimultaneousPredictionofAnti-enterococciActivities
andToxicologicalProfiles
463
Predictions performed by the mtk-QSBER model are
available in the Supplementary Information 4
(Suppl. Inf. 4) file. We need to emphasize that
predictions were realized against the most important
enterococci, i.e., Enterococcus faecalis and
Enterococcus faecium, which are the principal
bacteria of causing nosocomial infections.
According to the reports available for MIC
50
and
MIC
90
values in the webpage of Nabriva
Therapeutics, and the reference 41, BC-3781 may be
used to treat infections caused by Enterococcus
faecium, but not Enterococcus faecalis. Thus,
predictions made by the mtk-QSBER model,
confirm the experimental results. Also, in Suppl. Inf.
4, we performed predictions focused on the
toxicological profiles. According to the different
cutoff values of toxicities reported in Table 1, BC-
3781 can be a very safe antibacterial agent. Our
predictions help to explain why this pleuromutilin
derivative has undergone phase II clinical trials with
positive results. At the same time, we are
demonstrating that our mtk-QSBER model can be
used for virtual screening of toxicologically safe
anti-enterococci agents.
Figure 3: Chemical structure of the promising antibiotic
BC-3781.
4 CONCLUSIONS
Mt-QSAR approaches have emerged as novel and
powerful alternatives in the field of computer-aided
drug design, displaying very good performance for
the modeling of many different biological activities,
against diverse biological targets and experimental
conditions. In our work, we extended the mt-QSAR
concept by constructing an mtk-QSBER model that
allowed us to include not only biological (anti-
enterococci) activity data, but also, toxicological
profiles over several biological entities. Thus, our
mtk-QSBER model was developed to perform
simultaneous prediction of antibacterial activity
against bacteria of the genus Enterococcus spp. and
toxicity of compounds on laboratory animals and
human lymphocytes. The present mtk-QSBER
model confirms the idea that the use of mt-QSAR
methodologies permits to obtain more realistic and
accurate results. The performance of our mtk-
QSBER model, by classifying compounds as
positive or negative from a large and heterogeneous
database of compounds, and depending on dissimilar
measures of biological effects, targets, and
reliabilities of experimental conditions, permits its
use with one essential purpose: discovery of novel,
potent, versatile and safe anti-enterococci drug
candidates.
ACKNOWLEDGEMENTS
A. Speck-Planche acknowledges the Portuguese
Fundação para a Ciência e a Tecnologia (FCT) and
the European Social Found for financial support
(SFRH/BD/77690/2011).
REFERENCES
Balaban, A. T., 1982. Highly discriminating distance-
based topological index. Chemical Physics Letters.
89(5):399–404.
Brachman, P. S., Abrutyn, E., 2009. Bacterial Infections of
Humans: Epidemiology and Control, Springer
Science+Business Media, LLC. New York, NY.
Estrada, E., 1995. Edge adjacency relationship and a novel
topological index related to molecular volume.
Journal of Chemical Information and Computer
Sciences. 35:31–33.
Estrada, E., 2002. Physicochemical Interpretation of
Molecular Connectivity Indices. The Journal of
Physical Chemistry A. 106(39):9085–9091.
Estrada, E., Gutiérrez, Y., 2002-2004. MODESLAB, v1.5.
Santiago de Compostela.
Fisher, K., Phillips, C., 2009. The ecology, epidemiology
and virulence of Enterococcus. Microbiology. 155(Pt
6):1749-1757.
Flaherty, D., 2012. Immunology for Pharmacy, Mosby,
Elsevier Inc. St. Louis, Missouri.
Gaulton, A., Bellis, L. J., Bento, A. P., Chambers, J.,
Davies, M., Hersey, A., Light, Y., McGlinchey, S.,
Michalovich, D., Al-Lazikani, B., Overington, J. P.,
2012. ChEMBL: a large-scale bioactivity database for
drug discovery. Nucleic Acids Research. 40:D1100-
1107.
Hall, M., Frank, E., Holmes, G., Pfahringer, B.,
Reutemann, P., Witten, I. H., 1999-2013. WEKA
3.6.9. Waikato Environment for Knowledge Analysis.
Hamilton.
Hau, J., Schapiro, S. J., 2011. Handbook of Laboratory
Animal Science: Essential Principles and Practices,
CRC Press, Taylor & Francis Group, LLC. Boca
Raton, FL.
Hill, T., Lewicki, P., 2006. STATISTICS Methods and
IJCCI2013-InternationalJointConferenceonComputationalIntelligence
464

Applications. A Comprehensive Reference for Science,
Industry and Data Mining, StatSoft. Tulsa.
Kier, L. B., Hall, L. H., 1986. Molecular connectivity in
structure-activity analysis, Research Studies Press,
Wiley Letchworth, Hertfordshire, England, New York.
Munteanu, C. R., Magalhaes, A. L., Uriarte, E., Gonzalez-
Diaz, H., 2009. Multi-target QPDR classification
model for human breast and colon cancer-related
proteins using star graph topological indices. Journal
Theoretical Biology. 257(2):303-311.
Oprea, T., 2005. Chemoinformatics in Drug Discovery,
WILEY-VCH Verlag GmbH & Co. KGaA.
Weinheim.
Prado-Prado, F. J., Borges, F., Perez-Montoto, L. G.,
Gonzalez-Diaz, H., 2009. Multi-target spectral
moment: QSAR for antifungal drugs vs. different
fungi species. European Journal Medicinal Chemistry.
44(10):4051-4056.
Prado-Prado, F. J., Garcia-Mera, X., Gonzalez-Diaz, H.,
2010. Multi-target spectral moment QSAR versus
ANN for antiparasitic drugs against different parasite
species. Bioorganic and Medicinal Chemistry.
18(6):2225-2231.
Ryan, K. J., Ray, C. G., 2004. Sherris Medical
Microbiology McGraw Hill. Arizona.
Sader, H. S., Biedenbach, D. J., Paukner, S., Ivezic-
Schoenfeld, Z., Jones, R. N., 2012. Antimicrobial
activity of the investigational pleuromutilin compound
BC-3781 tested against Gram-positive organisms
commonly associated with acute bacterial skin and
skin structure infections. Antimicrobial Agents and
Chemotherapy. 56(3):1619-1623.
Speck-Planche, A., Kleandrova, V. V., 2012. In silico
design of multi-target inhibitors for C-C chemokine
receptors using substructural descriptors. Molecular
Diversity. 16(1):183-191.
Speck-Planche, A., Kleandrova, V. V., Cordeiro, M. N. D.
S., 2013. New insights toward the discovery of
antibacterial agents: Multi-tasking QSBER model for
the simultaneous prediction of anti-tuberculosis
activity and toxicological profiles of drugs. European
Journal Pharmaceutical Sciences. 48(4-5):812-818.
Speck-Planche, A., Kleandrova, V. V., Luan, F., Cordeiro,
M. N. D. S., 2012. Rational drug design for anti-
cancer chemotherapy: multi-target QSAR models for
the in silico discovery of anti-colorectal cancer agents.
Bioorganic and Medicinal Chemistry. 20(15):4848-
4855.
StatSoft, 2001. STATISTICA 6.0. Data analysis software
system.
Tenorio-Borroto, E., Penuelas Rivas, C. G., Vasquez
Chagoyan, J. C., Castanedo, N., Prado-Prado, F. J.,
Garcia-Mera, X., Gonzalez-Diaz, H., 2012. ANN
multiplexing model of drugs effect on macrophages;
theoretical and flow cytometry study on the
cytotoxicity of the anti-microbial drug G1 in spleen.
Bioorganic and Medicinal Chemistry. 20(20):6181-
6194.
Todeschini, R., Consonni, V., 2009. Molecular
Descriptors for Chemoinformatics, WILEY-VCH
Verlag GmbH & Co. KGaA. Weinheim.
ChemoinformaticsinDrugDesign.ArtificialNeuralNetworksforSimultaneousPredictionofAnti-enterococciActivities
andToxicologicalProfiles
465
