
Generalized Pythagoras Trees for Visualizing Hierarchies
Fabian Beck
1
, Michael Burch
1
, Tanja Munz
1
,
Lorenzo Di Silvestro
2
and Daniel Weiskopf
1
1
VISUS, University of Stuttgart, Stuttgart, Germany
2
Dipartimento di Matematica e Informatica, Universit
`
a di Catania, Catania, Italy
Keywords:
Hierarchy Visualization.
Abstract:
Pythagoras Trees are fractals that can be used to depict binary hierarchies. But this binary encoding is an
obstacle for visualizing hierarchical data such as file systems or phylogenetic trees, which branch into n sub-
hierarchies. Although any hierarchy can be modeled as a binary one by subsequently dividing n-ary branches
into a sequence of n − 1 binary branches, we follow a different strategy. In our approach extending Pythago-
ras Trees to arbitrarily branching trees, we only need a single visual element for an n-ary branch instead of
spreading the binary branches along a strand. Each vertex in the hierarchy is visualized as a rectangle sized
according to a metric. We analyze several visual parameters such as length, width, order, and color of the
nodes against the use of different metrics. The usefulness of our technique is illustrated by two case studies
visualizing directory structures and a large phylogenetic tree. We compare our approach with existing tree
diagrams and discuss questions of geometry, perception, readability, and aesthetics.
1 INTRODUCTION
Hierarchical data (i.e., trees) occurs in many applica-
tion domains, for instance, as results of a hierarchical
clustering algorithm, as files organized in directory
structures, or as species classified in a phylogenetic
tree. Providing an overview of possibly large and
deeply nested tree structures is one of the challenges
in information visualization. An appropriate visual-
ization technique should produce compact, readable,
and comprehensive diagrams, which ideally also look
aesthetically appealing and natural to the human eye.
A prominent visualization method are node-link
diagrams, which are often simply denoted as tree di-
agrams; layout and aesthetic criteria have been dis-
cussed (Reingold and Tilford, 1981; Wetherell and
Shannon, 1979). Although node-link diagrams are
intuitive and easy to draw, visual scalability and la-
beling often is an issue. An alternative, in particular
easing the labeling problem, are indented trees (Burch
et al., 2010) depicting the hierarchical structure by in-
dentation. Further, layered icicle plots (Kruskal and
Landwehr, 1983) stack boxes on top of each other for
encoding a hierarchy, but waste space by assigning
large areas to inner nodes on higher levels of the hier-
archy. The Treemap approach (Shneiderman, 1992),
which is applying the concept of nested boxes, pro-
duces space-efficient diagrams but complicates inter-
preting the hierarchical structure.
In this paper, we introduce Generalized Pythago-
ras Trees as an alternative to the above hierarchy visu-
alization techniques. It is based on Pythagoras Trees
(Bosman, 1957), a fractal technique showing a binary
hierarchy as branching squares (Figure 1, a); the frac-
tal approach is named after Pythagoras because every
branch creates a right triangle and the Pythagorean
theorem is applicable to the areas of the squares. We
extend this approach to n-arily branching structures
and use it for depicting information hierarchies (Fig-
ure 1, b). Instead of triangles, each recursive render-
ing step produces a convex polygonal shape where the
corners are placed on a semi circle. The size of the
created rectangles can be modified for encoding nu-
meric information such as the number of leaf nodes
of the respective subhierarchy (Figure 1, c).
We implemented the approach as an interactive
tool and demonstrate its usefulness by applying it to
large and deeply structured abstract hierarchy data
from two application domains: a file system orga-
nized in directories and the NCBI taxonomy, a phy-
logentic tree that structures the living organisms on
earth in a tree consisting of more than 300,000 ver-
tices. Furthermore, a comparison to existing hier-
archy visualization approaches provides first insights
17
Beck F., Burch M., Munz T., Di Silvestro L. and Weiskopf D..
Generalized Pythagoras Trees for Visualizing Hierarchies.
DOI: 10.5220/0004654500170028
In Proceedings of the 5th International Conference on Information Visualization Theory and Applications (IVAPP-2014), pages 17-28
ISBN: 978-989-758-005-5
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

(a) (b) (c)
Figure 1: Extending Pythagoras Trees for encoding information hierarchies: (a) traditional fractal approach; (b) Generalized
Pythagoras Tree applied to an n-ary information hierarchy; (c) additionally visualizing the number of leaves by the size of the
inner nodes.
into the unique characteristic of Generalized Pythago-
ras Trees: a higher visual variety leads to more dis-
tinguishable visualizations, the fractal origin of the
method supports identifying self-similar structures,
and the specific layout seems to be particularly suit-
able for visualizing deep hierarchies. Finally, the
created images are visually appealing as they show
analogies to natural tree and branching structures.
2 RELATED WORK
The visualization of hierarchical data is a central in-
formation visualization problem that has been stud-
ied for many years. Typical respresentations in-
clude node-link, stacking, nesting, indentation, or
fractal concepts as surveyed by J
¨
urgensmann and
Schulz (2010); Schulz (2011). Many variants of the
general concepts exist, for instance, radial (Battista
et al., 1999; Eades, 1992) and bubble layouts (Grivet
et al., 2004; Lin and Yen, 2007) of node-link dia-
grams, circular approaches for stacking techniques
(Andrews and Heidegger, 1998; Stasko and Zhang,
2000; Yang et al., 2003), or nested visualizations
based on Voronoi diagrams (Balzer et al., 2005; Nocaj
and Brandes, 2012).
Although many tree visualizations were proposed
in the past, none provides a generally applicable solu-
tion and solves all related issues. For example, node-
link diagrams clearly show the hierarchical structure
by using explicit links in a crossing-free layout. How-
ever, by showing the node-link diagram in the tradi-
tional fashion with the root vertex on top and leaves
at the bottom, much screen space stays unused at the
top while leaves densely agglomerate at the bottom.
Transforming the layout into a radial one distributes
the nodes more evenly, but makes comparisons of
subtrees more difficult. Node-link layouts of hierar-
chies have been studied in greater detail, for instance,
Burch et al. (2011) investigated visual task solution
strategies whereas McGuffin and Robert (2009) ana-
lyzed space-efficiency.
Indented representations of hierarchies are well-
known from explorable lists of files in file browsers.
Recently, Burch et al. (2010) investigated a variant
as a technique for representing large hierarchies as
an overview representation. Such a diagram scales
to very large and deep hierarchies and still shows
the hierarchical organization but not as clear as in
node-link diagrams. Layered icicle plots (Kruskal
and Landwehr, 1983), in contrast, use the concept of
stacking: the root vertex is placed on top and, analo-
gous to node-link diagrams, consumes much horizon-
tal space that is as large as all child nodes together.
Treemaps (Shneiderman, 1992), a space-filling
approach, are a prominent representative of nesting
techniques for encoding hierarchies. While proper-
ties of leaf nodes can be easily observed, a limitation
becomes apparent when one tries to explore the hi-
erarchical structure because it is difficult to retrieve
the exact hierarchical information from deeply nested
boxes: representatives of inner vertices are (nearly)
completely covered by descendants. Treemaps have
been extended to other layout techniques such as
Voronoi diagrams (Balzer et al., 2005; Nocaj and
Brandes, 2012) producing aesthetic diagrams that,
however, suffer from high runtime complexity.
Also, 3D approaches have been investigated, for
instance, in Cone Trees (Carri
`
ere and Kazman, 1995),
each hierarchy vertex is visually encoded as a cone
with the apex placed on the circle circumference of
the parent. Occlusion problems occur that are solved
by interactive features such as rotation. Botanical
Trees (Kleiberg et al., 2001), a further 3D approach,
imitate the aesthetics of natural trees but are restricted
to binary hierarchies, that is, n-ary hierarchies are
modeled as binary trees by the strand model of Holton
(1994); it becomes harder to detect the parent of a
node.
The term fractal was coined by Mandelbrot (1982)
and the class of those approaches has also been used
IVAPP2014-InternationalConferenceonInformationVisualizationTheoryandApplications
18

for hierarchy visualization due to their self-similarity
property (Koike, 1995; Koike and Yoshihara, 1993).
With OneZoom (Rosindell and Harmon, 2012), the
authors propose a fractal-based technique for visu-
alizing phylogenetic trees; however, n-ary branches
need to be visually translated into binary splits. De-
vroye and Kruszewski (1995) visualize random bi-
nary hierarchies with a fractal approach as botan-
ical trees; no additional metric value for the ver-
tices is taken into account; instead, they investigate
the Horton-Strahler number for computing the branch
thicknesses.
The goal of our work is to extend a fractal ap-
proach, which is closer to natural tree structures, to-
wards information visualization. This goal promises
embedding the idea of self-similarity and aesthet-
ics of fractals into hierarchy visualization. Central
prerequisite—and in this, our approach differs from
existing fractal approaches—is that n-ary branches
should be possible. With respect to information vi-
sualization, the approach targets at combining ad-
vantages of several existing techniques: a readable
and scalable representation, an efficient use of screen
space, and the flexibility for encoding additional in-
formation. A downside of the approach, however, is
that overlap may occur similar as in 3D techniques
(though it is a 2D representation)—only varying the
parameters of the visualization or using interaction al-
leviates this issue.
3 VISUALIZATION TECHNIQUE
Our general hierarchy visualization approach extends
the idea of Pythagoras Trees. Instead of basing the
branching of subtrees on right triangles, we exploit
convex polygons with edges on the circumference of
a semi circle.
3.1 Data Model
We model a hierarchy as a directed graph H = (V, E)
where V = {v
1
, . . . , v
k
} denotes the finite set of k
vertices and E ⊂ V × V the finite set of edges, i.e.,
parent–child relationships. One vertex is the desig-
nated root vertex and is the only vertex without an
incoming edge; all other vertices have an in-degree of
one. We allow arbitrary hierarchies, that is, the out-
degree of the vertices is not restricted. A maximum
branching factor n ∈ N of H can be computed as the
maximum out-degree of all v ∈ V . For an arbitrary
vertex v ∈ V , H
v
denotes the subhierarchy having v as
root vertex; | H
v
| is the number of vertices included
in the H
v
(including v). The depth of a vertex v
0
in
(a) (b)
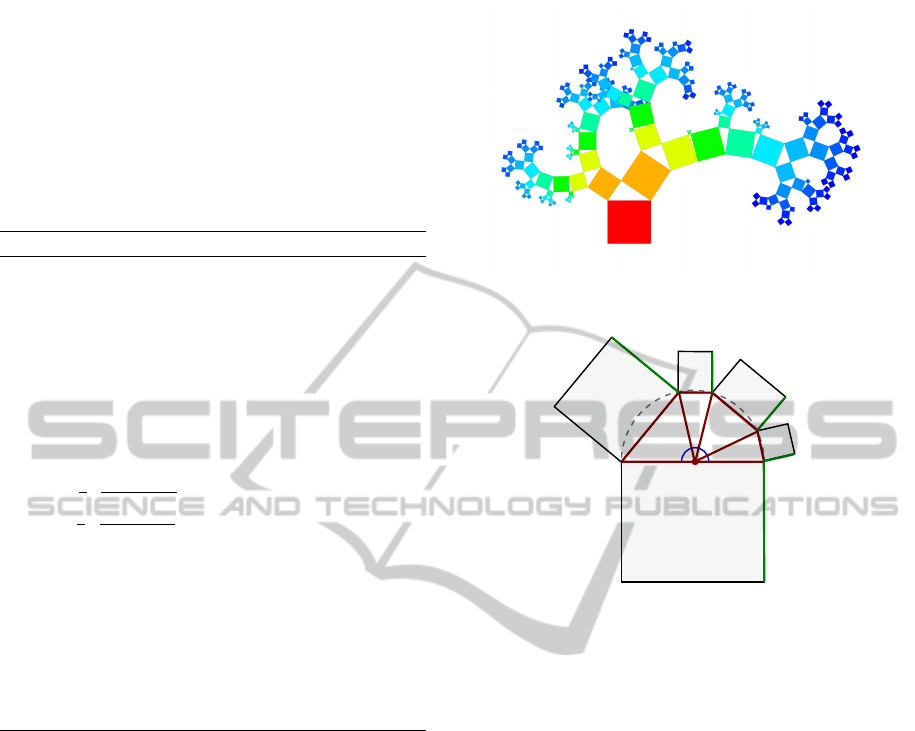
Figure 2: Illustration of the traditional Pythagoras Tree ap-
proach: (a) a single binary branch; (b) recursively applied
branching step.
H
v
is the number of vertices on the path through the
hierarchy from v to v
0
. We allow positive weights to
be attached to each vertex of the hierarchy v ∈ V rep-
resenting metric values such as sizes. We model them
as a function w : V → R. The weight w(v) ∈ R
+
of an
inner vertex v does not necessarily need to be the sum
of its children, but can be.
3.2 Traditional Pythagoras Tree
The Pythagoras Tree is a fractal approach describing
a recursive procedure of drawing squares. In that, it
was initially not intended to encode information, but
its tree structure easily allows representing binary hi-
erarchies: each square represents a vertex of the hier-
archy; the recursive generation follows the structure
of the hierarchy and ends at the leaves.
Drawing a fractal Pythagoras Tree starts with
drawing a square of side length c. Then, two smaller
squares are attached at one side of the square—
usually, at the top—according to the procedure illus-
trated in Figure 2 (a): Then, a right triangle with an-
gles α and β where α + β =
π
2
is drawn using the
side of the square as hypotenuse, which also becomes
a diameter of the circumcircle of the triangle. The
two legs of the triangle are completed to squares hav-
ing side lengths a and b. In the right triangle, the
Pythagorean theorem a
2
+b
2
= c
2
holds, i.e., the sum
of the areas of the squares over the legs is equal to
the area of the square over the hypotenuse. Applying
this procedure recursively to the new squares as de-
picted for the next step in Figure 2 (b) creates a frac-
tal Pythagoras Tree (the recursion is only stopped for
practical reasons at some depth). The angles α and β
can be set to a constant value or be varied according to
some procedural pattern. Figure 1 (a) provides an ex-
ample of a fractal Pythagoras Tree where α = β =
π
4
.
Transforming the fractal approach into an infor-
mation visualization technique, the squares are inter-
preted as representatives of vertices of the hierarchy,
GeneralizedPythagorasTreesforVisualizingHierarchies
19
called nodes. As a consequence, the fractal encodes
a complete binary hierarchy, the recursion depth be-
ing the depth of the hierarchy. If the generated image
should represent a binary hierarchy that is not com-
pletely filled to a certain depth, the recursion has to
stop earlier for the respective subtrees. If the hier-
archy is weighted as specified in the data model, the
weights can be visually encoded by adjusting the sizes
of the squares, i.e., the corresponding angles α and β.
Algorithm 1: Pythagoras Tree.
PythagorasTree(H
v
, S):
// H
v
: binary hierarchy
// S: representative square S = (c, ∆s, θ)
// c = (x
c
, y
c
): center
// ∆s: length of a side
// θ: slope angle
drawSquare(S); // draw square for current root vertex
if | H
v
|> 1 then
// v
1
and v
2
: children of H
v
α :=
π
2
·
w(v
2
)
w(v
1
)+w(v
2
)
;
β :=
π
2
·
w(v
1
)
w(v
1
)+w(v
2
)
;
∆s
1
:= ∆s · sin β;
∆s
2
:= ∆s · sin α;
c
1
:= ComputeCenterLe f t(c, ∆s, ∆s
1
, );
c
2
:= ComputeCenterRight(c, ∆s, ∆s
2
);
S
1
:= (c
1
, ∆s
1
, θ + α);
S
2
:= (c
2
, ∆s
2
, θ − β);
PythagorasTree(H
v
1
, S
1
); // draw subhierarchy H
v
1
PythagorasTree(H
v
2
, S
2
); // draw subhierarchy H
v
2
end if
Algorithm 1 describes in greater detail how an ar-
bitrary binary hierarchy (i.e., a hierarchy where each
vertex either has an out-degree of 2 or 0) can be recur-
sively transformed into a Pythagoras Tree visualiza-
tion. It is initiated by calling PythagorasTree(H
v
, S):
where H
v
= (V, E) is a binary hierarchy and S =
(c, ∆s, θ) is the initial square with center c, length of
the sides ∆s, and slope angle θ. The recursive pro-
cedure first draws square S and proceeds if the cur-
rent hierarchy still contains more than a single node.
Then, encoding the node weights in the size of the
squares, the angles α and β are computed according
to the normalized weight of the node opposed to the
angle. The angles form the basis for further com-
puting the parameters of the two new rectangles S
1
and S
2
. The drawing procedure is finally continued
by recursively calling PythagorasTree(H
v
1
, S
1
) and
PythagorasTree(H
v
2
, S
2
) for the two children v
1
and
v
2
of the current root vertex.
When, for instance, using the number of leaf ver-
tices as the weight of each vertex, the algorithm pro-
duces visualizations such as Figure 3 that encodes a
Figure 3: Random binary hierarchy visualized as a
Pythagoras Tree that encodes the number of leaves in the
size of the nodes.
v
4
v
3
v
2
v
1
α
1
α
2
α
3
α
4
R
Δx
1
Δy
1
Δx
2
Δy
2
Δx
3
Δy
3
Δx
4
Δy
4
R
1
R
2
R
3
R
4
v
Δy
Δx
Figure 4: Polygonal split of Generalized Pythagoras Trees
creating an n-ary branch.
random binary hierarchy. Like the fractal approach,
the visualization algorithm still produces overlap of
subtrees that, however, becomes rarer through sparser
hierarchies.
3.3 Generalized Pythagoras Tree
The Generalized Pythagoras Tree, as introduced in the
following, can be used for visualizing arbitrary hier-
archies, that are hierarchies allowing n-ary branches.
Right triangles are replaced by convex polygons shar-
ing the same circumcircle; the former hypotenuse of
the triangle becomes the longest side of the triangle.
For increasing the visual flexibility of the approach,
squares are exchanged for general rectangles.
Figure 4 illustrates an n-ary branch, showing the
polygon and its circumcircle. The polygon is split
into a fan of isosceles triangles using the center of
the circumcircle as splitting point. While the number
of rectangles is specified by the degree of the repre-
sented branch, the angles and lengths can be modified
to encode further information. In particular, we have
two degrees of freedom:
• Width function w
x
: V → R
+
of rectangles: Sim-
ilar to binary hierarchies, the width ∆x
i
of a rect-
angle R
i
can be changed, here, by modifying the
IVAPP2014-InternationalConferenceonInformationVisualizationTheoryandApplications
20

corresponding angle α
i
accordingly. The angle α
i
should reflect weight w
x
(v
i
) of a vertex v
i
in rela-
tion to the weight of its siblings:
α
i
:= π ·
w
x
(v
i
)
∑
n
j=1
w
x
(v
j
)
.
The width of the rectangle is ∆x
i
:= ∆x · sin
α
i
2
where ∆x is the width of the parent node.
• Length stretch function w
y
of rectangles: Anal-
ogously, the length ∆y
i
of the rectangle R
i
can be
varied. This length, in contrast to the width ∆x
i
,
does not underly any restrictions such as the size
of a cirumcircle. Nevertheless, we formulate the
length dependent on the length of the parent ∆y
and the relative width sin
α
i
2
in order to consider
the visual context (otherwise, it would be diffi-
cult to define appropriate metrics not producing
degenerated visualizations): the length of the rect-
angle is ∆y
i
:= w
y
(v
i
) · ∆y · sin
α
i
2
.
Algorithm 2: Generalized Pythagoras Tree.
GeneralizedPythagorasTree(H
v
, R):
// H
v
: hierarchy branching into n ∈ N
0
subhierarchies
H
v
1
, . .. , H
v
n
// R: representative rectangle R = (c, ∆x, ∆y, θ)
// c = (x
c
, y
c
): center
// ∆x, ∆y: width and length
// θ: slope angle
drawRectangle(R); // draw rectangle for parent vertex
for all H
v
i
do
α
i
:= π ·
w
x
(v
i
)
∑
n
j=1
w
x
(v
j
)
;
∆x
i
:= ∆x · sin
α
i
2
;
∆y
i
:= w
y
(v
i
) · ∆y · sin
α
i
2
;
c
i
:=ComputeCenter(c, ∆x, ∆y, (α
1
, . .. , α
i−1
), ∆x
i
, ∆y
i
);
θ
i
:=ComputeSlope(θ, (α
1
, . .. , α
i
));
R
i
:= (c
i
, ∆x
i
, ∆y
i
, θ
i
);
GeneralizedPythagorasTree(H
v
i
, R
i
);
end for
Algorithm 2 extends Algorithm1 and describes the
generation of Generalized Pythagoras Tree visualiza-
tions. Again, it is a recursive procedure and is initial-
ized by calling GeneralizedPythagorasTree(H
v
, R)
where H
v
= (V, E) is an arbitrary hierarchy and R =
(c, ∆x, ∆y, θ) represents the initial rectangle that, in
contrast to the previous case, has a width ∆x and
a length ∆y. For an n-ary branching hierarchy H
v
with root vertex v, the algorithm first draws the re-
spective rectangle before all children v
1
, . . . , v
n
are
handled: for each child v
i
, the computation of an-
gle α
i
forms the basis for deriving the width ∆x
i
and length ∆y
i
of the respective rectangle R
i
as de-
scribed above. Furthermore, the center and slope
Figure 5: Generalized Pythagoras Trees showing n-ary hi-
erarchy using a constant width and length stretch function.
of the new rectangle need to be retrieved. Finally,
GeneralizedPythagorasTree(H
v
i
, R
i
) can be recur-
sively applied to subhierarchy H
v
i
having rectangle R
i
as root node.
Figure 5 shows a sample visualization created
with the algorithm. For this initial image width func-
tion w
x
is set to a constant value and the length stretch
function w
y
is defined as 1. As a consequence, the
nodes are squares again, equally sized for each branch
but n-arily branching. An example with a similar
configuration can be found in Figure 1 (a); the same
dataset is shown in Figure 1 (b) applying the num-
ber of leaf nodes as the width function w
x
. Further
configurations are discussed more systematically be-
low. The discussion also includes the usage of color,
which, in all figures referenced so far, visualizes the
depth of the nodes. Furthermore, the order of rectan-
gles can be modified and has an impact on the lay-
out; in the generalized approach, we have a higher
degree of freedom (n! possibilities) than in the stan-
dard Pythagoras Trees where only a flipping between
two angles can be applied.
3.4 Excursus: Fractal Dimension
The fractal dimension is typically used as a complex-
ity measure for fractals. Looking back to the origin of
the Generalized Pythagoras Tree visualization and in-
terpreting it as a fractal approach, the extended fractal
approach can be characterized by this dimension. To
this end, however, not an information hierarchy can
be encoded, but the approach needs to be applied for
infinite n-arily branching structures; for simplification
we do not consider scaling of rectangles. The follow-
ing analysis shows that the fractal dimension, which
is 2 for traditional Pythagoras Tree fractals, asymptot-
ically decreases to 1 for a branching factor approach-
ing infinity.
Any fractal can be characterized by its fractal di-
mension D ∈ R that is defined as a relation between
GeneralizedPythagorasTreesforVisualizingHierarchies
21
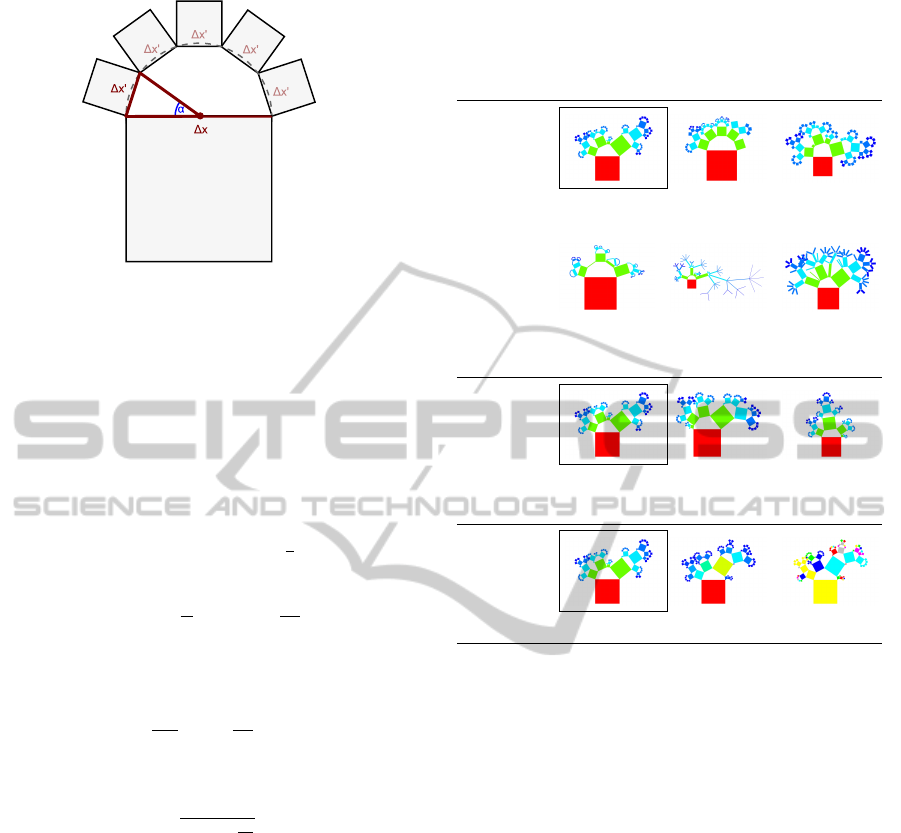
Figure 6: Illustrating the fractal dimension of an n-ary
branching hierarchy by showing the splitting into equally
sized angles.
the branching factor n and the scaling factor r given
by D = −log
r
n. In our scenario, we have to first com-
pute the scaling factor r depending on the branching
factor n. Figure 6 illustrates the following formulas
and shows an n-ary branch.
First of all, the n-ary branch creates a convex poly-
gon, which is split into isosceles triangles as described
before. Since all rectangles have the same width, the
angle at the tip of the triangle is α =
π
n
. The width of
the rectangle then is
∆x
0
= ∆x · sin
α
2
= ∆x · sin
π
2n
.
Relating the size of the square to the original square,
the scaling factor can be derived as follows:
r =
∆x
0
∆x
= sin
π
2n
.
The fractal dimension finally is
D
n
= −
log n
log sin
π
2n
.
This result confirms D
2
= 2 (traditional binary
branches) and shows that the fractal dimension is ap-
proaching 1 for increasing n, i.e.,
lim
n→∞
D
n
= 1 .
3.5 Visual Parameters
The visualization approach has been described pre-
cisely but still has some degrees of freedom that shall
be explored in the following. For example, the size of
the rectangles can be varied, the order of the subhier-
archies in a branch is not restricted, or the coloring of
the nodes is open for variation. These parameters help
optimizing the layout and support the visualization by
extra information in form of weights assigned to each
node. For illustrating the effect, Table 1 shows the
Table 1: Exploring different parameter settings such as size,
order, and color of rectangles for a sample dataset; framed
images represent the default setting and are equivalent; the
number of leaf nodes is applied as weight.
size
(squares) (S1) sides: weight (S2) sides: equal
size
(S3) sides:
weight; enlarged
circle
size
(rectangles) (S4) width: equal
size; length:
weight
(S5) width: equal
size; area: weight
(S6) width:
weight; area:
weight
order
(O1) external (O2) ascending
weight
(O3) maximum
weight in the
center
color
(C1) depth (C2) weight (C3) category
same random hierarchy (75 nodes; maximum depth
of 5) in different parameter settings. As a weight,
the number of leaf nodes is applied; but the metric is
interchangeable, for instance, by the number of sub-
nodes, the depth of the subtree, or a domain-specific
weight. One setting (Table 1, S1 = O1 = C1), which
seemed to work most universally in our experience, is
selected as default and applied in all following figures
of the paper if not indicated otherwise.
3.5.1 Size
Already for the traditional Pythagoras Tree approach,
rectangles can be split in uniform size or non-uniform
size. For the generalized approach, we define a width
function as well as a length function (Section 3.3).
When employing the same metric for both, all nodes
are represented as squares. Table 1 (S1) uses the num-
ber of leaf nodes as the common metric, which seems
to be a good default selection because more space is
assigned to larger subtrees. In contrast, when all sub-
nodes are assigned the same size (i.e. a constant func-
tion is employed), small subtrees become overrepre-
sented as depicted in Table 1 (S2). A variant of the
approach, which is shown in Table 1 (S3), extends the
IVAPP2014-InternationalConferenceonInformationVisualizationTheoryandApplications
22

approach from using semi circles to larger sectors of
a circle.
Inserting different functions for width and length
further increases the flexibility—nodes are no longer
squares, but differently shaped rectangles. For in-
stance, Table 1 (S4) encodes the number of leaf nodes
in the height and applies a constant value to the width.
When defining the length function relative to the (con-
stant) width so that the area of the rectangle is propor-
tional to the number of leaves, those leaf nodes are
emphasized as depicted in Table 1 (S5). A similar
variant shown in Table 1 (S6) has a constant length
and chooses the width accordingly for encoding the
number of leaf nodes in the area.
3.5.2 Order
The subnodes of an inner node of a hierarchy are vi-
sualized as an ordered list. While, for some applica-
tions, there exist a specific, externally defined order,
many other scenarios do not dictate a specific order.
In case of the latter, the subnodes can be sorted ac-
cording to a metric, which again is the number of leaf
nodes in this example. The sorting criterion mainly
influences the direction in which the diagram is grow-
ing but also influences overlapping effects. Often the
external order, at least in case it is random or inde-
pendent of size, creates quite balanced views as de-
picted in Table 1 (O1). When, for instance, applying
an ascending order, the image like the one shown in
Table 1 (O2) grows to the right. More symmetric visu-
alizations such as in Table 1 (O3) are generated when
placing the vertices with the larger size in the center.
3.5.3 Color
The areas of the rectangular nodes can be filled with
color for encoding some extra information. Selecting
the color on a color scale according to the depth of
the node in the hierarchy helps comparing the depth
of subtrees: for instance, in Table 1 (C1) this encod-
ing reveals that the leftmost main subtree, though be-
ing shorter, is as deep as the rightmost one. Alterna-
tively, the weight of a node can be encoded in color
like shown in Table 1 (C2), which, however, is more
suitable if the size of the node not already encodes
the weight. If categories of vertices are available, also
these categories can be color-coded by discrete colors
as depicted in Table 1 (C3).
3.6 Analogy to Node-Link Diagrams
Though being derived from a fractal approach, Gen-
eralized Pythagoras Trees can be adapted—without
changing the position of nodes—to become variants
(a) (b) (c)
Figure 7: Relationship between Generalized Pythagoras
Trees and node-link diagrams: (a) Generalized Pythagoras
Tree; (b) Generalized Pythagoras Tree and analogous node-
link diagram; (c) analogous node-link diagram.
of node-link diagrams. An analogous diagram can
be created as illustrated in Figure 7 by connecting
the circle centers of the semi circles of branches by
lines. The circle centers become the nodes, the lines
become the links of the resulting node-link diagram.
Like the subtrees of a Generalized Pythagoras Tree
might overlap, the analogous node-link drawing is not
guaranteed to be free of edge crossings. We prefer the
Pythagoras variant over the analogous node-link vari-
ant because it uses the available screen space more
efficiently (which is important, for instance, for color
coding) and shows the width of a node explicitly.
4 CASE STUDIES
To illustrate the usefulness of our Generalized
Pythagoras Tree visualization, we applied it to two
datasets from different application domains—file sys-
tems with file sizes as well as the NCBI taxonomy that
classifies species. In these case studies we demon-
strate different parameter settings and also show how
interactive features can be applied for exploration.
4.1 File System Hierarchy
While the approach can be applied to any directory
structure, we decided to demonstrate this use case
by reading in the file structure of an early version of
this particular paper. Since we use L
A
T
E
X for writing,
the paper directory contains multiple text files includ-
ing temporary files as well as a list of images. Also
included are supplementary documents and a script
used for creating exemplary random information hier-
archies. All in all, the directory structure contains 139
vertices (7 directories and 132 files) having a maxi-
mum depth of 4 and a maximum branching factor of
38 (figures directory). Figure 8 shows two visualiza-
tions of this directory structure employing different
parameter settings.
In Figure 8 (a), we applied the default settings siz-
ing the vertices in relation to the number of leaf nodes
and using color for encoding depth. The image shows
that, among the main directories, the figures directory
GeneralizedPythagorasTreesforVisualizingHierarchies
23

(a) (b)
Figure 8: Directory hierarchy of this paper on the file sys-
tem: (a) size based on the number of leaf nodes with color-
coded depth information; (b) size encoding the file and di-
rectory sizes with color-coded file types.
contains by far the most leaf nodes (94) and itself is
split into three further directories, which include the
images needed for the three more complex figures and
tables of this paper: canis (Figure 9), parameters (Ta-
ble 1), and samples (Table 2). Additionally, figures
also directly includes a number of images, which are
needed for the other figures. The only other directory
containing a reasonable number of files is the hierar-
chy generator folder; besides the generator script it
contains a number of generated sample datasets.
Customizing the parameters of the visualization
for the use case of investigating file systems, we as-
signed the file size to the size of the vertices (directory
sizes are the sum of the contained file sizes). More-
over, the file type is encoded in the color of the vertex
(category coding) a legend providing the color–type
assignments; directories are encoded in the color of
the dominating file type of the contained files. The re-
sulting visualization as depicted in Figure 8 (b) shows
that the figures directory is also one of the largest
main directories, but there exist other files and direc-
tories that also consume reasonable space such as the
additional material directory. Comparing the size of
the main PDF document to the images directory, it can
be observed that not all image files contained in the di-
rectory are integrated into the paper because the paper
is smaller than the images directory. The color-coded
file types reveal that the most frequently occurring
type are PNG files, not only in the images directory
but also in general. The hierarchy generator direc-
tory mostly includes TRE files (Newick format), but
is dominated with respect to size by two TXT files (an
alternative hierarchy format not as space-efficient).
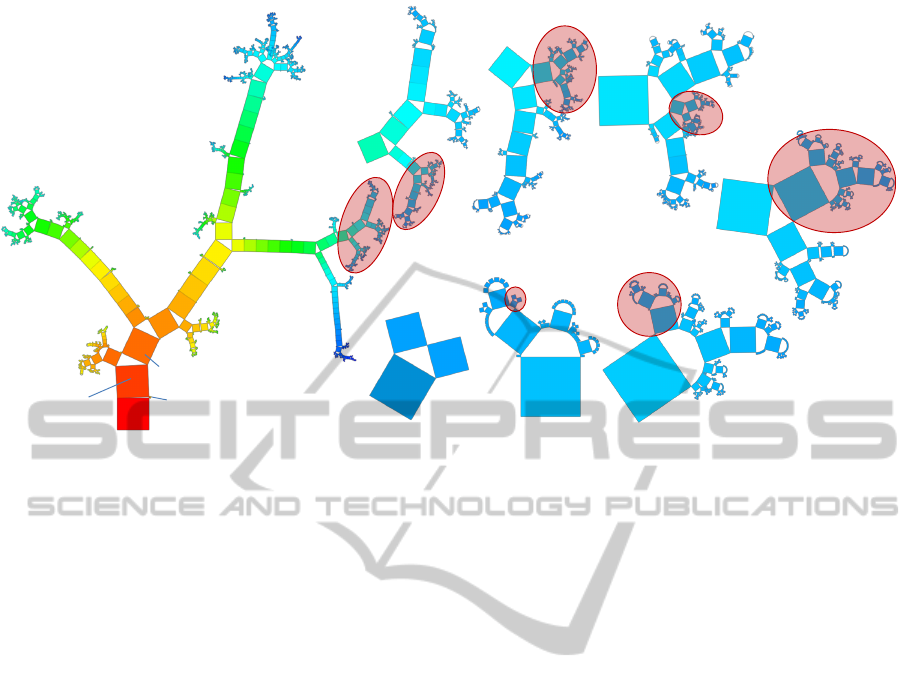
4.2 Phylogenetic Tree
Moreover, our approach is tested on a hierarchical
dataset commonly used by the biology and bioinfor-
matics communities. The taxonomy here used has
been developed by NCBI and contains the names
of all organisms that are represented in its genetic
database (Benson et al., 2010). The specific dataset
encoding the taxonomy contains 324,276 vertices
(60,585 classes and 263,691 species) and has a max-
imum depth of 42. The Generalized Pythagoras Tree
visualization applied to this dataset (Figure 9 I) cre-
ates a readable overview visualization of the very
complex and large hierarchical structure. The vertices
of the tree have different sizes according to the num-
ber of leaves of their subtrees. Each inner vertex rep-
resents a class of species and it is easy to point out
the class that contains more species. The root node
is an artificial class of the taxonomy that contains ev-
ery species for which a DNA sequence or a protein is
stored in the NCBI digital archive.
At the first level of the tree (see Figure 9 I), a big
node represents cellular organisms and further nodes
the Viruses, Viroids, unclassified species, and others
(this information can be retrieved by using the ge-
ometric zoom). Selecting nodes and retrieving ad-
ditional information facilitate the exploration of the
tree. For instance, the biggest node at level 2 is the
Eukaryota class, which includes all organisms whose
cells contain a membrane-separated nucleus in which
DNA is aggregated in chromosomes; it still contains
177,258 of the 263,691 species.
Besides gaining an overview of the main branches
of the taxonomy, the visualization tool allows for an-
alyzing subsets of the hierarchy down to the level of
individual species by applying semantic zooming. As
a concrete example, we demonstrate the exploration
process in the right part of Figure 9; in each step we
selected the subtree of the highlighted node (red cir-
cle): Figure 9 II shows the Amniota class, which be-
longs to the tetrapoda vertebrata taxis (four-limbed
animals with backbones or spinal columns). In the
next steps (Figure 9 III-V), we followed interesting
branches until we reach the Carnivora class in Fig-
ure 9 V, which denotes meat-eating organisms; the
subtree contains 301 species. From here, it is simple
to proceed the exploration towards a well-known an-
imal, such as the common dog, defined as Canis Fa-
miliaris, by zooming in the subtrees of Caniformia,
literally “dog shaped” (Figure 9 VI), then through
Canidae, the family of dogs (Figure 9 VII) with 45
species, and finally Canis Familiaris.
5 DISCUSSION
The introduced technique for representing hierar-
chical structures is discussed by taking existing
other hierarchy visualization approaches into account.
IVAPP2014-InternationalConferenceonInformationVisualizationTheoryandApplications
24
II. Amniota
III. Eutheria
IV. Laurasiatheria
V. Carnivora
VI. Caniformia
VII. Canidae VIII. Canis Familiaris
I. NCBI Taxonomy
Eukaryota
cellular
organisms
others
Figure 9: NCBI taxonomy hierarchically classifying species; rectangles sizes indicate the number of species in a subtree,
color encodes the depth; an example for exploring the taxonomy by semantic zooming is provided.
We applied different standard hierarchy visualization
techniques to a number of randomly generated artifi-
cial datasets. The results are listed in Table 2. Each
column represents a different data set with some char-
acteristic feature: a binary hierarchy with a branch-
ing factor of 2, a deep hierarchy with many levels, a
flat hierarchy with a high maximum branching factor,
a degenerated hierarchy that grows linearly in depth
with the number of nodes, a symmetric hierarchy hav-
ing two identical subtrees, and a self-similar hierarchy
following the same pattern at each level. The rows
show standard visualization techniques in comparison
to Generalized Pythagoras Trees. Though the graph-
ics can only act as previews in a printed version of the
paper, they are included in high resolution and are ex-
plorable in a digital version. The following analysis
considers multiple levels of abstraction from geome-
try and perception to readability and aesthetics.
5.1 Geometry and Perception
Hierarchy visualizations aim at showing containment
relationships between nodes and their descendants.
Considering Gestalt theory (Wertheimer, 1923), dif-
ferent approaches exist for visually encoding rela-
tionships: for instance, node-link diagrams use con-
nectedness to express containment, while Treemaps
are based on common region for showing that several
nodes belong to the same parent. In contrast, Gen-
eralized Pythagoras Trees do neither directly draw a
line between the nodes nor nest one node into the
other, but they draw rectangles of decreasing size onto
an imaginary curve. The human reader automatically
connects the rectangles on the curve, which is denoted
as the law of continuation. In all hierarchy visual-
ization approaches shown in Table 2, proximity also
plays a certain role (i.e., related nodes are placed next
to each other) but should not be overinterpreted (i.e.,
nodes placed next to each other are not necessarily
related).
In node-link diagrams, indented tree diagrams, or
icicle plots, each level in the hierarchy creates an-
other layer in the visualization. As a consequence,
the amount of (vertical) space available for a layer is
reduced when adding further levels. In Generalized
Pythagoras Trees, however, there are no global lay-
ers for levels of nodes: adding a level only produces
a kind of local layer that is arranged on a semi cir-
cle. With respect to this characteristic, Generalized
Pythagoras Trees are similar to Treemaps, which nei-
ther have global layers but split the area of a node for
introducing the next level.
Like in icicle plots and Treemaps, larger areas are
used to encode the nodes in Generalized Pythagoras
Trees. This makes it easier to use color for encod-
ing some metrics (such as the hierarchy level) in the
nodes because colors are easier to perceive for larger
areas (Ware, 2004) (Color for Labeling). In contrast
to Treemaps (and complete icicle plots), Generalized
Pythogoras Trees do not create space-filling images.
Areas, however, might overlap, which is discussed in
detail below.
Comparing the images shown in Table 2 with re-
spect to uniqueness, Generalized Pythagoras Trees
GeneralizedPythagorasTreesforVisualizingHierarchies
25
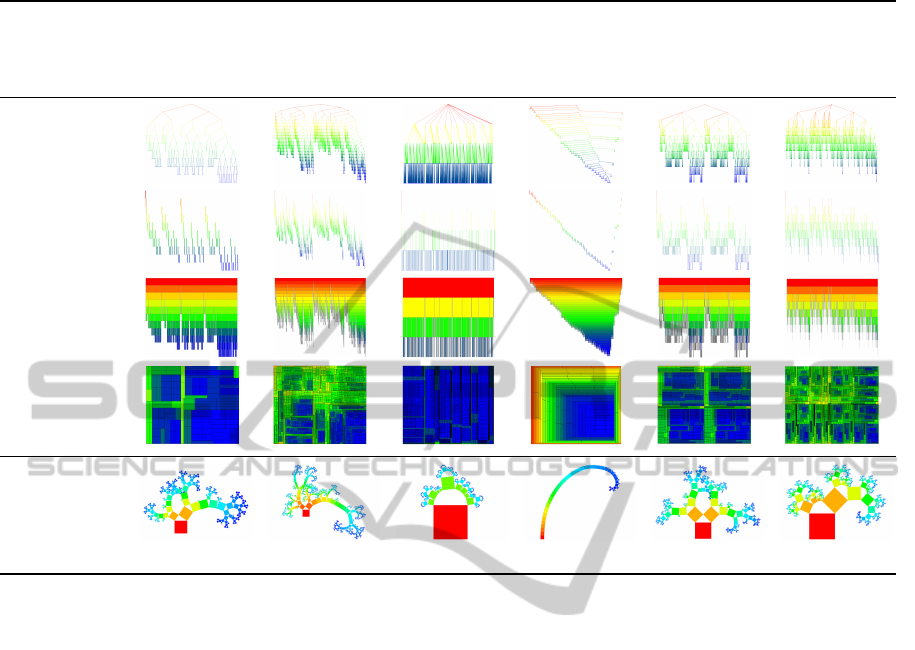
Table 2: Comparison of hierarchy visualization approaches for representatives of a selected set of hierarchy classes.
binary hierarchy deep hierarchy flat hierarchy degenerated
hierarchy
symmetric
hierarchy
self-similar
hierarchy
node degree of 2 high number of
hierarchy levels (25)
high maximum node
degree (20)
linearly growing depth two equivalent
subtrees
self similar tree
structure
node-link
indented tree
icicle plot
Treemap
Generalized
Pythagoras Tree
show a high visual variety: not only the subtrees vary
in size, they are also rotated. Only the splitting ap-
proach in Treemaps creates similarly varying images,
however, just with respect to texture but not shape.
A positive effect of a high visual variety is that the
different datasets can be distinguished more easily—
the visualization acts as a fingerprint. Together with
the fractal roots of the approach, the uniqueness helps
detect self-similar structures: Table 2 (last column)
shows a tree having a self-similar structure, which is
generated according to the same recursive, determin-
istic procedure for every node; the self-similar prop-
erty of the hierarchy is best detectable in the Gener-
alized Pythagoras Trees because every part of the tree
is just a rotated version of the complete tree.
5.2 Readability and Scalability
A hierarchy visualization is readable if the users are
able to efficiently retrieve the original hierarchical
data from it and easily observe higher-level charac-
teristics. However, readability is also related to vi-
sual scalability, which means preserving readability
for larger datasets. While, for smaller datasets, the
exact information is usually recognizable in any hi-
erarchy visualization, the depicted information often
becomes too detailed when increasing the scale of the
dataset. The visualization approach, hence, needs to
use the available screen space efficiently and has to
focus on the most important information.
Generalized Pythagoras Trees clearly emphasize
the higher-level nodes of the tree (i.e., the root node
and its immediate descendants): most of the area that
is filled by the visualization is consumed by these
higher-level nodes, which can be easily perceived be-
cause surrounded by whitespace. Lower-level nodes
and leaf nodes, however, become very small and are
not visible. But the visualization allows for sizing
the nodes according to their importance by using the
number of leaf nodes as a metric as done in Table 2.
Node-link diagrams, indented trees, and icicle plots
are similar in their focus on the higher-level nodes;
as well, lower-level nodes become difficult to discern
because of lack of horizontal space. Since the ver-
tical space assigned to each level does not become
smaller in these visualizations, it is easier to retrieve
the maximum depth of a subtree. Treemaps focus on
leaf nodes and show largely different characteristics.
The ability of a visualization technique to display
also large datasets in a readable way considerably
widens its area of application. As shown in the case
study, Generalized Pythagoras Trees can be used for
browsing large hierarchies such as the NCBI taxon-
omy. While it is possible to interactively explore large
IVAPP2014-InternationalConferenceonInformationVisualizationTheoryandApplications
26

hierarchies in a similar way with the other paradigms
listed in Table 2, Generalized Pythagoras Trees show
some characteristic scalability advantages: for specif-
ically deep hierarchies such as the one in the sec-
ond column of Table 2, it adaptively expands into
the direction of the deepest subtree, here in spiral
shape. Comparing it to the other approaches, deep
subtrees are still readable in surprising detail. In con-
trast for flat hierarchies, which have a specifically
high branching factor, Generalized Pythagoras Trees
do not seem to be as suitable: the size of the nodes
decreases too fast which constrains readability.
For a degenerated hierarchy (Table 2, fourth col-
umn), which grows linearly in depth with the number
of nodes, Generalized Pythagoras Trees create an id-
iosyncratic but readable visualization, similar as it is
the case for the other visualization approaches. Also
a symmetry in a hierarchy such as two identical sub-
trees (Table 2, fifth column) can be detected: the iden-
tical tree creates the same image, which is rotated in
contrast to the other approaches, where it is moved
but not rotated.
A problem limiting the readability of Generalized
Pythagoras Trees is that, depending on the visualized
hierarchy, subtrees might overlap. The other visual-
ization approaches do not share this problem; only
Treemaps also employ a form of overplotting: inner
nodes are overplotted by its direct descendants. While
Treemaps use overplotting systematically, overlap
only occurs occasionally in Generalized Pythagoras
Trees and is unwanted. A simple way to circumvent
the problem using the interactive tool is selecting the
subset of the tree that is overdrawn by another. Also,
reordering the nodes or adapting the parameters of the
algorithm could alleviate the problem.
5.3 Aesthetics
Fractals often show similarities to natural structures
such as trees, leaves, ferns, clouds, coastlines, or
mountains (Peitgen and Saupe, 1988). Among the
images shown in Table 2, the Generalized Pythagoras
Trees clearly show the highest similarity to natural
tree and branching structures. Since, according to
the biophilia hypothesis, humans are drawn towards
every form of life (Wilson, 1984), this similarity
suggests that Generalized Pythagoras Trees might
be considered as being specifically aesthetic. Also
the property of self-similarity that is partly preserved
when generalizing Pythagoras Trees supports aesthet-
ics: “fractal images are usually complex, however, the
propriety of self-similarity makes these images easier
to process, which gives an explanation to why we
usually find fractal images beautiful.” (Machado and
Cardoso, 1998)
6 CONCLUSIONS AND FUTURE
WORK
In this paper, we introduced an extension of Pythago-
ras Tree fractals with the goal of using these for vi-
sualizing information hierarchies. Instead of depict-
ing only binary trees, we generalize the approach
to arbitrarily branching hierarchy structures. An al-
gorithm for generating these Generalized Pythagoras
Trees was introduced and the fractal characteristics of
the new approach were reported. A set of parame-
ters allows for customizing the approach and creating
a variety of visualizations. In particular, metrics can
be visualized for the nodes. The approach was imple-
mented in an interactive tool. A case study demon-
strates the utility of the approach for analyzing large
hierarchy datasets. The theoretical comparison of
Generalized Pythagoras Trees to other hierarchy visu-
alization paradigms, on the one hand, suggested that
the novel approach is capable of visualizing various
features of hierarchies in a readable way comparably
to previous approaches and, on the other hand, might
reveal unique characteristics of the approach such as
an increased distinguishability of the generated im-
ages and detectabiltiy of self-similar structures. Fur-
ther, the approach may have advantages for visualiz-
ing deep hierarchies and provides natural aesthetics.
An open research questions is how the overplot-
ting problem of the approach can be solved efficiently
and how the assumed advantages can be leveraged in
practical application. Moreover, formal user studies
have to be conducted to further explore the character-
istics of the approach.
ACKNOWLEDGMENTS
We would like to thank Kay Nieselt, University of
T
¨
ubingen, for providing the NCBI taxonomy dataset.
REFERENCES
Andrews, K. and Heidegger, H. (1998). Information slices:
Visualising and exploring large hierarchies using cas-
cading, semicircular disks. In Proceedings of IEEE
Symposium on Information Visualization, pages 9–11.
Balzer, M., Deussen, O., and Lewerentz, C. (2005). Voronoi
treemaps for the visualization of software metrics.
GeneralizedPythagorasTreesforVisualizingHierarchies
27

In Proceedings of Software Visualization, pages 165–
172.
Battista, G. D., Eades, P., Tamassia, R., and Tollis, I. G.
(1999). Graph Drawing: Algorithms for the Visual-
ization of Graphs. Prentice-Hall.
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell,
J., and Sayers, E. W. (2010). Genbank. Nucleic Acids
Research, 38(suppl 1):D46–D51.
Bosman, A. E. (1957). Het wondere onderzoekingsveld der
vlakke meetkunde. Breda, N.V. Uitgeversmaatschappij
Parcival.
Burch, M., Konevtsova, N., Heinrich, J., H
¨
oferlin, M., and
Weiskopf, D. (2011). Evaluation of traditional, or-
thogonal, and radial tree diagrams by an eye tracking
study. IEEE Transactions on Visualization and Com-
puter Graphics, 17(12):2440–2448.
Burch, M., Raschke, M., and Weiskopf, D. (2010). Indented
Pixel Tree Plots. In Proceedings of International Sym-
posium on Visual Computing, pages 338–349.
Carri
`
ere, S. J. and Kazman, R. (1995). Research report: In-
teracting with huge hierarchies: beyond cone trees. In
Proceedings of Information Visualization, pages 74–
81.
Devroye, L. and Kruszewski, P. (1995). The botanical
beauty of random binary trees. In Proceedings of
Graph Drawing, pages 166–177.
Eades, P. (1992). Drawing free trees. Bulletin of the Institute
for Combinatorics and its Applications, 5:10–36.
Grivet, S., Auber, D., Domenger, J., and Melanc¸on, G.
(2004). Bubble tree drawing algorithm. In Proceed-
ings of International Conference on Computer Vision
and Graphics, pages 633–641.
Holton, M. (1994). Strands, gravity, and botanical tree
imaginery. Computer Graphics Forum, 13(1):57–67.
J
¨
urgensmann, S. and Schulz, H.-J. (2010). A visual survey
of tree visualization. IEEE Visweek 2010 Posters.
Kleiberg, E., van de Wetering, H., and van Wijk, J. J.
(2001). Botanical visualization of huge hierarchies. In
Proceedings of Information Visualization, pages 87–
94.
Koike, H. (1995). Generalized fractal views: A
fractal-based method for controlling information dis-
play. ACM Transactions on Information Systems,
13(3):305–324.
Koike, H. and Yoshihara, H. (1993). Fractal approaches for
visualizing huge hierarchies. In Proceedings of Visual
Languages, pages 55–60.
Kruskal, J. and Landwehr, J. (1983). Icicle plots: Better dis-
plays for hierarchical clustering. The American Statis-
tician, 37(2):162–168.
Lin, C. C. and Yen, H. C. (2007). On balloon drawings
of rooted trees. Graph Algorithms and Applications,
11(2):431–452.
Machado, P. and Cardoso, A. (1998). Computing aesthetics.
In Advances in Artificial Intelligence, volume 1515 of
Lecture Notes in Computer Science, pages 219–228.
Springer Berlin Heidelberg.
Mandelbrot, B. (1982). The Fractal Geometry of Nature.
W.H. Freeman and Company. New York.
McGuffin, M. and Robert, J. (2009). Quantifying the space-
efficiency of 2D graphical representations of trees. In-
formation Visualization, 9(2):115–140.
Nocaj, A. and Brandes, U. (2012). Computing
Voronoi Treemaps: Faster, simpler, and resolution-
independent. Computer Graphics Forum, 31(3):855–
864.
Peitgen, H.-O. and Saupe, D., editors (1988). Science of
Fractal Images. Springer-Verlag.
Reingold, E. and Tilford, J. (1981). Tidier drawings of trees.
IEEE Transactions on Software Engineering, 7:223–
228.
Rosindell, J. and Harmon, L. (2012). OneZoom: A fractal
explorer for the tree of life. PLOS Biology, 10(10).
Schulz, H.-J. (2011). Treevis.net: A tree visualization ref-
erence. IEEE Computer Graphics and Applications,
31(6):11–15.
Shneiderman, B. (1992). Tree visualization with tree-maps:
2-D space-filling approach. ACM Transactions on
Graphics, 11(1):92–99.
Stasko, J. T. and Zhang, E. (2000). Focus+context display
and navigation techniques for enhancing radial, space-
filling hierarchy visualizations. In Proceedings of the
IEEE Symposium on Information Visualization, pages
57–65.
Ware, C. (2004). Information Visualization, Second Edi-
tion: Perception for Design (Interactive Technolo-
gies). Morgan Kaufmann, 2nd edition.
Wertheimer, M. (1923). Untersuchungen zur Lehre von der
Gestalt. II. Psychological Research, 4(1):301–350.
Wetherell, C. and Shannon, A. (1979). Tidy drawings of
trees. IEEE Transactions on Software Engineering,
5(5):514–520.
Wilson, E. O. (1984). Biophilia. Harvard University Press.
Yang, J., Ward, M. O., Rundensteiner, E. A., and Patro, A.
(2003). InterRing: A visual interface for navigating
and manipulating hierarchies. Information Visualiza-
tion, 2(1):16–30.
IVAPP2014-InternationalConferenceonInformationVisualizationTheoryandApplications
28
