
An Intelligent Inference Engine using Ontology based Clinical
Pathways for Diagnosis and Management of Diabetes
Shreelakshmi G. M., Kavya A. K. Alse, Ananthakrishna Thantri, Krupesha D. and Srinivas A.
PES Institute of Technology,100 Feet Ring Road, BSK III Stage, Bangalore 560085, India
Keywords: Clinical Pathways, Web Ontology Language (OWL), Resource Description Framework (RDF).
Abstract: ‘Clinical pathways’ for Diabetes Management has attracted the attention of researchers in the last decade.
Ontologies have been in use to represent knowledge pertaining to clinical pathways and to arrive at critical
patient-specific decisions. This paper proposes an ontological framework to represent the diabetes related
data. The main contribution of the paper is in developing an inference model that helps a General
Practitioner (GP) to arrive at the most appropriate clinical pathway for a patient specific condition. The
mobile application developed for this purpose makes it very useful for a medical practitioner in a remote
rural location to follow a systematic process to arrive at patient specific decisions, based on the Ontological
inferences received from the remote server.
1 INTRODUCTION
The whole world and especially developing
countries like India are facing an alarming situation
with an increasing number of Diabetes patients.
India is termed the “Diabetes Capital” of the world
and by 2030, nearly 10% of the population will be
affected by the disease. The serious situation is
compounded by the fact that a sizable population
(nearly 52%) is unaware of the existence of high
blood sugar levels which can lead to Type-II
Diabetes, mainly due to lack of education. Juvenile
onset of Diabetes is also on the rise and if it is not
diagnosed in the early stages, it can lead to
complications in the later stages of life. Diabetes
can lead to serious life threatening diseases such as
coronary heart disease and stroke. High levels of
sugar in the blood accelerate the filtering of blood by
the kidneys. The additional work on the filters can
result in leaks in the filters and useful proteins are
lost in the urine. This results in deterioration of
kidney functionality. If left unattended, Diabetes
can cause Diabetic Retinopathy which may result in
complete loss of sight. In general, almost every part
of the body can get affected by the disease.
Clinical Pathways (CP) are structured, multi-
disciplinary plans of care designed to support the
implementation of clinical guidelines and protocols.
They provide detailed guidance for each stage in the
management of a patient (treatment, intervention
etc.), with a specific condition over a given time
period, and include progress and outcome details.
For diagnosis, prescription and patient management,
clinical pathways provide various alternatives that
are derived for a spectrum of patient conditions and
it is for the medical expert to decide on a specific
pathway for a patient specific condition.
Diabetic Ontology is the knowledge
representation pertaining to Diabetes and it helps to
develop models for the entities such as patient,
hospital, and doctor etc. to interact with the
knowledge provider (Ahmed, 2011). Use of an
ontology based model enables separation of domain
knowledge from operational logic. It is a significant
merit of ontological approach over the traditional if-
then rule based approach. It not only captures and
expresses the structure and semantics of the domain
knowledge, but also enables developing software
agents which support decision making. There has
been a significant interest in developing ontology for
clinical pathways in the last decade (Islam, Freytag,
and Shankar, 2012; Ahmed, 2011; Lin, 2011;
Nimmagadda, Nimmagadda and Dreher, 2011;
Chen, and Hadzic, 2010; Chen, Bau, and Huang,
2010; McGarry, Garfield, and Wermter, 2007).
Robust models have been built to represent
knowledge in the ontological frame work.
An OWL/SWRL enabled ontological model that
aids in the development of a software tool to provide
patient specific reminders, advise and action items
143
G. M. S., A. K. Alse K., Thantri A., D. K. and A. S..
An Intelligent Inference Engine using Ontology based Clinical Pathways for Diagnosis and Management of Diabetes.
DOI: 10.5220/0004707301430150
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2014), pages 143-150
ISBN: 978-989-758-010-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

with regard to the prevention of diabetic foot has
been developed by Chammas et al (Chammas,
2013). The paper uses a reasoning technique using
the semantics and ontological matching. Kurozumi,
et al (Kurozumi, 2013) present a Fuzzy Mark-up
Language (FML) based Japanese Diet Assessment
System. The tool helps a patient to manage his
healthy diet level making a judicious choice of a
range of available food items. Based on pre-defined
ontologies including ingredients of food items, the
Fuzzy Inference Mechanism suggests the dietary
health constitution on a personalized basis for a one-
day meal.
The research work proposed by Ahmed (Ahmad,
2011) focuses on developing ontologies in the OWL
language with Protégé as the modelling tool. It
suggests a Type-2 diabetes framework that uses a
three-layer model to develop the self-management
framework. Lee, Wang, and Hagras, (Lee, 2010)
propose a fuzzy ontology for a personalized
diabetic-diet recommendation in their work. The
authors apply the Type-2 Fuzzy set based intelligent
ontological agent for recommending the right type of
Diet-planning for a patient specific condition.
Personalized diabetic care is very essential to suit
the specific patient situation. The patient details such
as health information, pharmaceutical care, diet care,
sports care have been aggregated and diabetic care
ontology has been built by Chen, Su, and Chang,
(Chen, 2010). For a new patient needing an advice
of care, an ontology querying process has been built
in to the system in this research.
In their research on ontology based Decision
making, Chen, Chi, & Bau, (Chen, 2011) have
proposed a robust model to represent the diabetic
knowledge and a “Multiple Criteria Decision
Making (MCDM)” has been developed to compute
the right medication for a patient. The scheme uses
entropy to compute the patient data history and is
integrated with the knowledge ontology to arrive at
the personalized prescriptions. The research carried
out by Alhazbi, et al (Alhazbi, 2012) uses ontology
to represent food items and their nutritional
information. In addition to assisting patients to log
their glucose levels over a period of time via a
mobile device to a remote server, the application
helps patients to manage their food consumption
through the ontological knowledge representation.
A very interesting alternative technique of
arriving at inferences based on action rules has been
proposed by Hajja, et al (Hajja, 2013). The action
rules that describe possible state transitions during
the operational life cycle of a system with respect to
a decision attribute are extracted from the object
driven and temporal systems. The support and
confidence computed for the system will influence
the strength of action rules and the final inference.
The technique has been applied to the speech
disorder problem in children called “Hyponasality”.
A majority of Indian population is rural. This
underserved population entirely depends on the
services of a General Medical Practitioner (GP).
GPs in rural areas treat patients with varied
medical problems including Diabetes, in the absence
of specialists. With advances in technology and
medical practices, there would be newer methods
and techniques in diagnosis, treatment and
management of diabetes that the remotely located
GP may not be aware of. Also, there are many
newly introduced drugs which are to be administered
with caution, taking in to account, their side-effects.
This paper focuses on developing an Ontological
framework to represent clinical pathways for
Diabetes management. The framework encompasses
the representation of all possible clinical pathways in
terms of classes and objects in an ontology
developed using Protégé platform. An intelligent
inferencing mechanism has been developed that uses
the ontological knowledge to arrive at the optimal
clinical pathway for a patient specific condition. A
mobile application has been developed and it can be
used by a remotely located medical practitioner to
guide him on the optimal pathway. The mobile
device takes the patient specific conditions as input
attributes. It contacts the remotely located server that
has all the clinical pathways represented in the
ontological framework. The remotely located server
uses the inference engine to arrive at the most
optimal patient-specific pathway. The remote mobile
device with the GP will be able to receive this
pathway prescription and will be able to guide the
doctor in suitably advising the patient (in terms of
diagnosis, treatment, interventions etc.). It is very
important to note that the proposed system is not
meant to replace a medical expert in any way. This
will only aid in making sure that all the known
criteria are taken in to consideration, during
diagnosis, treatment and management of the disease.
2 ONTOLOGICAL FRAMEWORK
The basic principal that governs the proposed
scheme is as follows: The Medical practitioner in a
remote location provides the primary symptoms and
related details of his patient to a mobile device. This
part of the functionality that takes effect on the
mobile is termed the “Client Side Activity”
HEALTHINF2014-InternationalConferenceonHealthInformatics
144
developed using the Android. The processed queries
are passed on to the server which contains
all the server side functions in the form of Servlets.
The results of the Ontological processing and
decision making using the servlets are passed on to
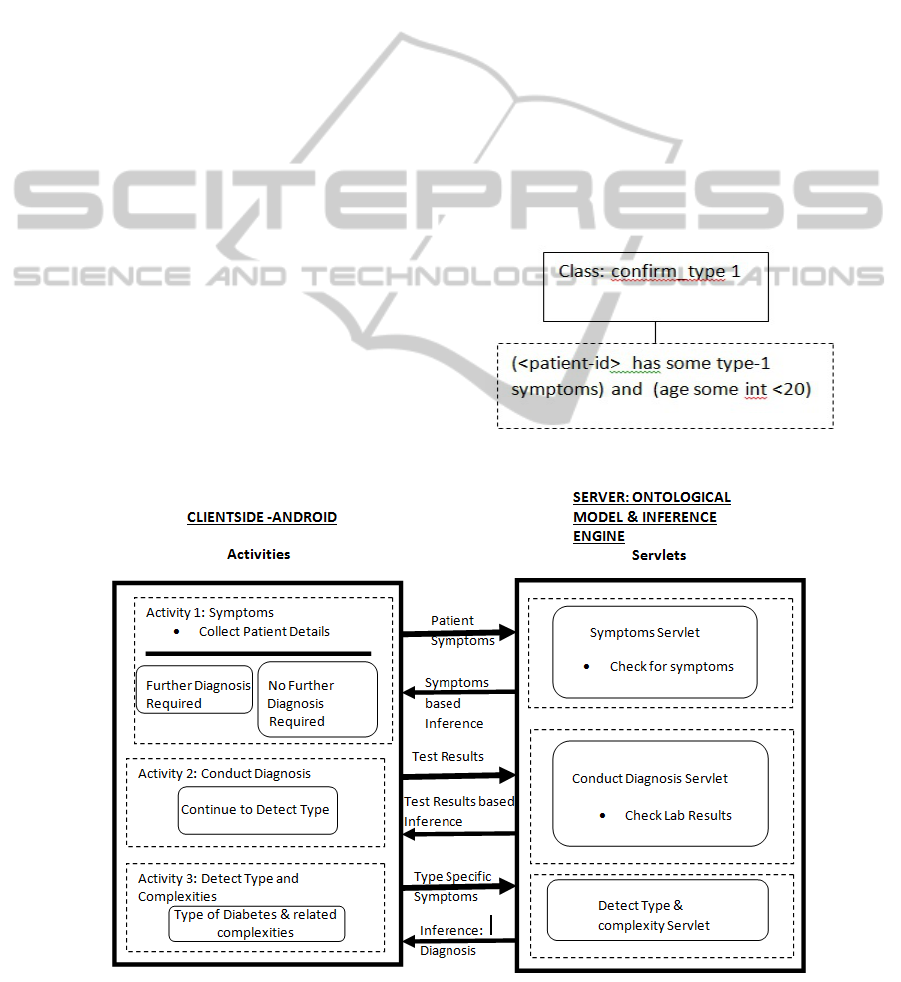
the Client. Figure 1 represents the generic model
proposed in this research work.
In Figure 1, the overall framework has two main
entities: The Client Side Android Activities and the
Server Side Ontological Model. The client prepares
the patient data as a string and sends it to
the server. At the server, the string is converted to
RDF triples. These triples in conjunction with the
restrictions defined in the OntModel together
facilitate the inferencing process.
2.1 Server Side Ontological Model
The Open Source Ontology Tool, Protégé is used to
represent the various entities of the model. The
model consists of Classes and Subclasses and
resources represent the attributes of a class. An
example for class representation is shown in Figure
2. “Properties” connect two resources. For example,
the property ‘has’ connects the two resources
‘patient_id’ and ‘excessive_thirst’ as depicted
below:
(Patient_id ) has (excessive_thirst)
The Class Hierarchy is represented in the ‘OWL’
file. The OWL file is loaded to the system memory
as “OntModel”. Thus the OntModel has the entire
Class Hierarchy together with the corresponding
resources and more importantly, the restrictions
accompanying the classes that will later be used in
the inference engine. The server side “Servlets”
implement the functionality for the inference engine.
In the present work, the three types of Servlets
considered are ‘Symptoms Servlet’, ‘Conduct
Diagnosis Servlet’ and ‘Detect Type & Complexity
Servlet’. These three servlets receive requests from
the client and use Jena APIs to act on the
Ontologies.
The three activities that get influenced by the
Jena APIs are:
Load Ontology
Parse Ontology
Parse the Result of Reasoner
Figure 2: Class and Restrictions.
Figure 1: System Model and Framework.
AnIntelligentInferenceEngineusingOntologybasedClinicalPathwaysforDiagnosisandManagementofDiabetes
145
2.2 Client Side Activity
When the patient related details are provided at the
client side by a mobile device, the data is sent to the
server as a string. This string is converted to triples
and added to the OntModel. When the server
responds with a resolution, the client displays this
and initiates further queries for the next level
resolution at the server end.
3 DESIGN
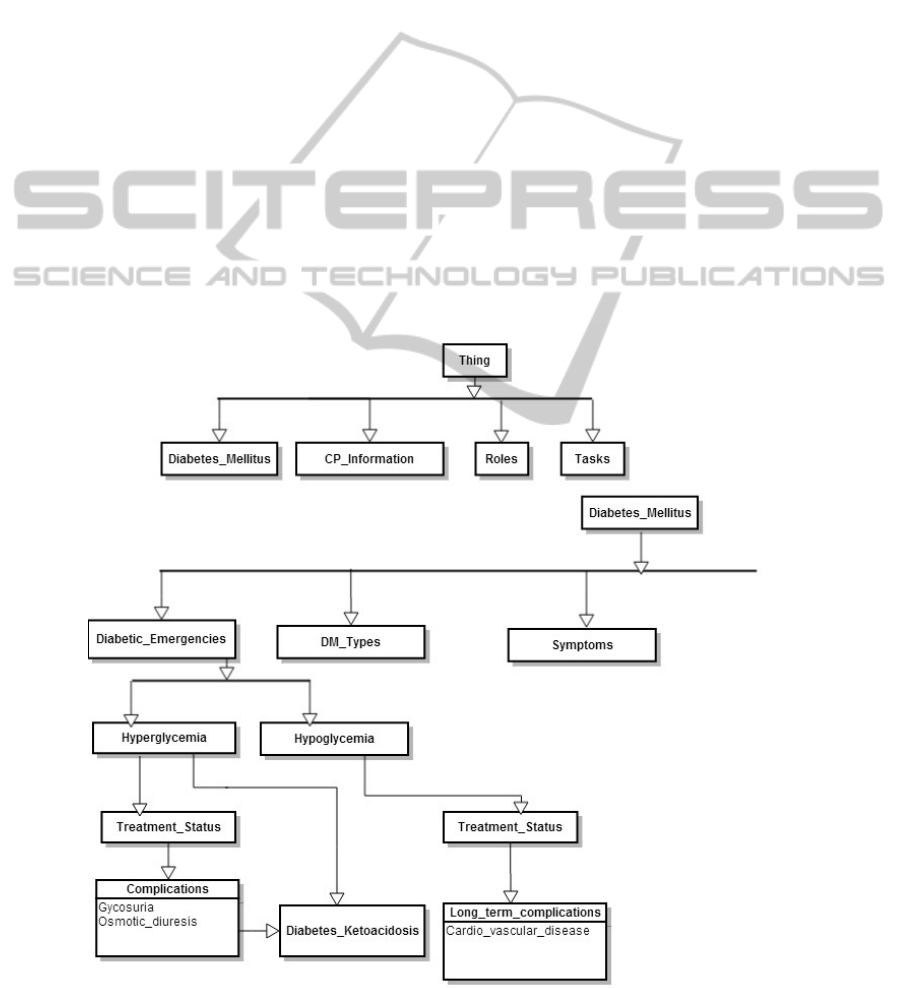
3.1 Class Hierarchy
The ontological representation in its entirety is
designed as a set of classes and sub-classes. Each of
these classes and subclasses has resources and
restrictions that will specifically be used in the
inference engine. An example of Class Hierarchy is
shown in Figure 3.
In the model developed in this paper, 90 classes
have been designed and incorporated in to Protégé.
The complete knowledge representation would
need a few hundreds of classes to realistically
represent most patient scenarios. This will
progressively be addressed in future designs, in
consultation with medical experts. In Figure 4, a
sample snapshot of one of the possible clinical
pathways to diagnose a patient as Type-1 or Type-2
is provided. The level to which the class hierarchy
is defined and the inference engine is developed,
will determine the number of clinical pathways
present in the system developed.
3.2 Clinical Pathways
The complete tree structure of the Class Hierarchy
together with the class restrictions has been used to
represent various possible clinical pathways. Each
pathway represents a particular scenario from any
one of Diagnosis, Treatment or Management.
3.3 Inference Engine
For a patient specific condition, the clinical and
other related parameters will be sent from the
Figure 3: Class Hierarchy.
HEALTHINF2014-InternationalConferenceonHealthInformatics
146
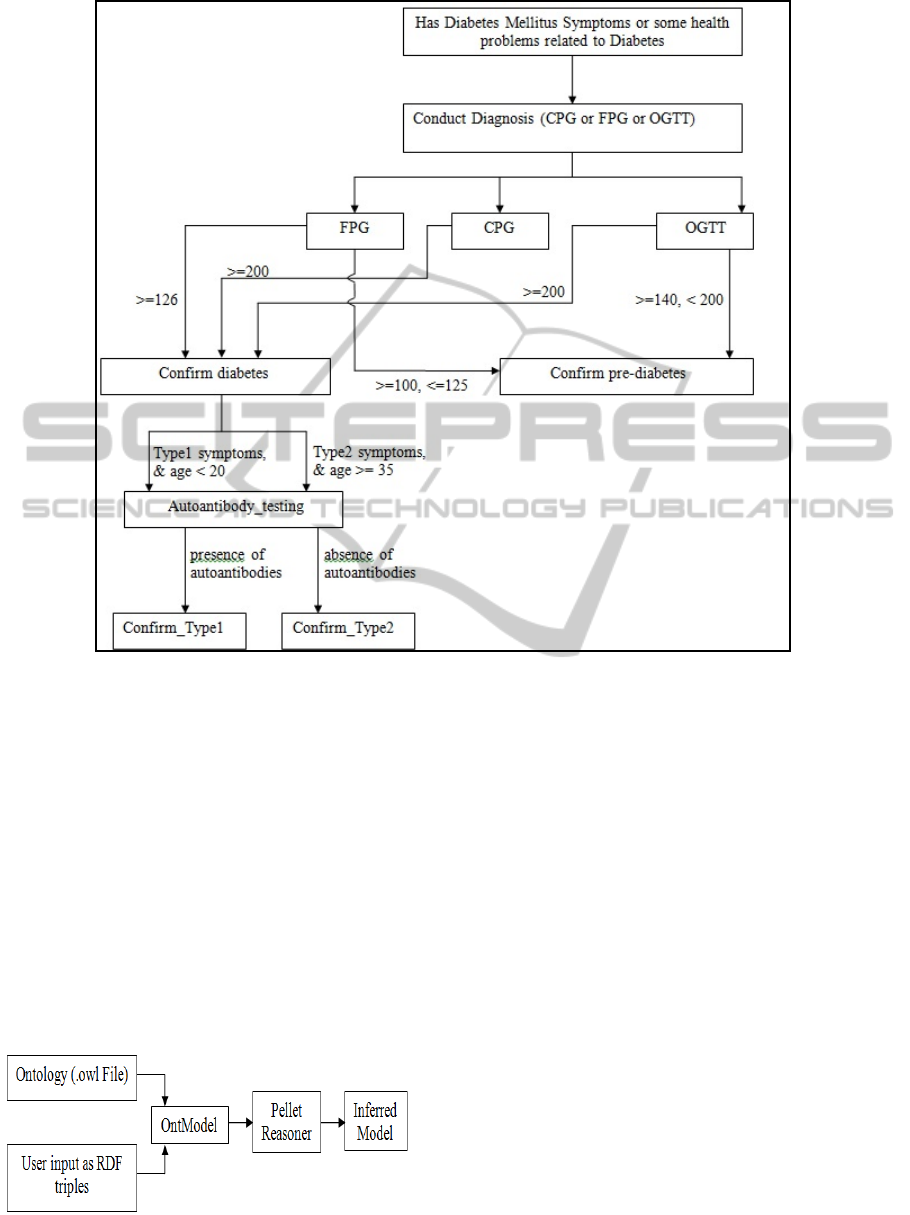
Figure 4: ‘Diagnosis’ Clinical Pathway Scenario.
mobile device to the server. The server performs the
inferencing activity via the inference engine and
sends out the output. Figure 5 shows the inferencing
process. The inferencing process adopts the
following steps using Jena APIs: (i) Loads the
ontology (from owl file) into an OntModel. (ii) Adds
patient data entered by the medico in the form of
RDF Triples (iii) The Ontmodel acts as an input to
the Pellet Reasoner. The Reasoner generates the
Inferred Model (based on classes, properties and
more importantly, the restrictions). (iv) The inferred
model is parsed using the Jena APIs and the results
are sent in the form of strings to the mobile device.
Figure 5: Inferencing Mechanism.
4 RESULTS AND DISCUSSION
The generic model brought out in Figure 1 is now
applied to two patient specific scenarios at the client
end and the results are discussed here.
Case Study 1: Diabetes Type Detection
(i) The patient data as indicated in Figure 6 is passed
on to the server. The server side program
(SymptomsServlet.java) runs the reasoner and gets
the inferred results using the restriction:
(has some DM_Symptoms) or (has some
Health_problems_related_to_diabetes)
This restriction aids the reasoner to infer that the
patient has to undergo further clinical tests and this
decision is passed on to the client.
(ii) In Figure 6, when “Conduct Diagnosis” is
selected, this is passed on to the “Conduct
Diagnosis” servlet and with CPG test result value as
221 (Figure 7).
The following restriction on the server side will
help to arrive at a decision to detect if the patient is
AnIntelligentInferenceEngineusingOntologybasedClinicalPathwaysforDiagnosisandManagementofDiabetes
147
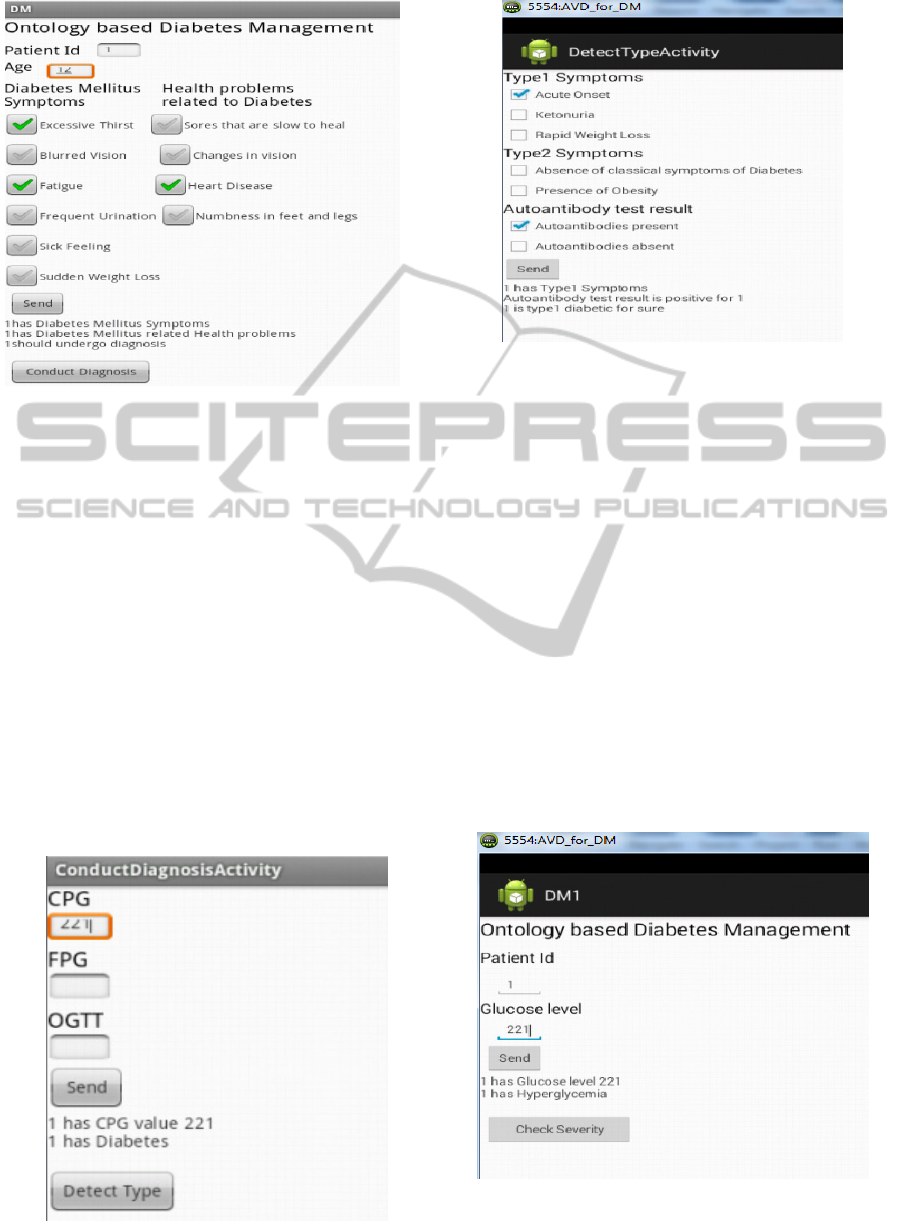
Figure 6: Patient Input Data on the Mobile.
to be classified to have “Diabetes Mellitus” based on
Casual Plasma Glucose (CPG), Fasting Plasma
Glucose (FPG) and Oral Glucose Tolerance Test
(OGTT):
(hasCPGValue some int[>= 200]) or
(hasFPGValue some int[>= 126]) or
(hasOGTTValue some int[>= 200])
The client module provides further clinical details
and the final diagnosis is provided by the servlet as
shown in Figure 8, classifying the patent as “Type-
1”.
The Patient is tested for the presence of
Autoantibodies. If Autoantibodies are present, it is
confirmed that the patient is Type 1 diabetic. The
following restriction helps in this decision.
(has some Type1_symptoms) and
(autoantibodies value "present")
Figure 7: Conduct Diagnosis Activity for Type Diagnosis.
Figure 8: Inference Engine Classifying the Patient as
“Type-1”.
Case Study 2: Test for Diabetic Ketoacidosis
The patient specific data is sent to the server as
shown in Figure 9. In this study, since the glucose
levels are abnormally high, the “Detect Type &
Complexity” servlet uses the Reasoner to infer that
the patient is Hyperglycemic.
Once it is confirmed that the patient is
Hyperglycemic, severity of Hyperglycemia is
decided depending on the treatment history as shown
in Figure 10, using the restrictions defined in the
ontology.
Figure 10 shows the test for presence of ketones
in blood or urine which helps in the diagnosis of
Diabetic Ketoacidosis. The following restriction is
used:
(has some Ketone_Testing)
Figure 9: Test for Hyperglycemia.
HEALTHINF2014-InternationalConferenceonHealthInformatics
148

Figure 10: Test for severity of Hyperglycemia.
Figure 11: Inference on Diabetic Ketoacidosis.
The Inference engine uses the presence of Ketones
in the blood to infer the patient condition as Diabetic
Ketoacidosis, as shown in Figure 11.
5 CONCLUSIONS
This paper proposes an Ontology based model to
represent various possible clinical pathways for
differing patient conditions. The classes, properties
and restrictions together help the inference model to
arrive at appropriate decisions with regard to
Diagnosis, Treatment or the Management. The main
contribution of the paper is the development of an
intelligent inference engine for decision making
which can be very useful in remote rural areas where
an expert Diabetologist is not easily accessible. The
system will be highly useful for General
Practitioners to take appropriate reasoning based
decisions, taking in to account all the recent
developments in diagnosis and treatment methods.
The pathways can be further extended to include
complications like nephropathy, retinopathy,
neuropathy etc. Future works also include providing
personalized (patient-specific) management in the
form of diet, exercise and drugs (based on
parameters like patient’s family history, disease
history etc). In collaboration with Diabetologists of
the institute medical school, the authors propose to
add a wide variety of classes to take in to account,
the domain experts’ decision process. Work is in
progress to achieve these goals.
ACKNOWLEDGEMENTS
The authors wish to thank Visvesvaraya
Technological University for their research grant to
carry out this work.
REFERENCES
Ahmed, A. S. (2011, July). Towards an Online Diabetes
Type II Self Management System: Ontology
Framework. In Computational Intelligence,
Communication Systems and Networks (CICSyN),
2011 Third International Conference on (pp. 37-41).
IEEE.
Alhazbi, S., Alkhateeb, M., Abdi, A., Janahi, A., &
Daradkeh, G. (2012, April). Mobile application for
diabetes control in Qatar. In Computing Technology
and Information Management (ICCM), 2012 8th
International Conference on (Vol. 2, pp. 763-766).
IEEE.
Chammas, N., Juric, R., Koay, N., Gurupur, V., & Suh,. C.
(2013, January). Towards a Software Tool for Raising
Awareness of Diabetic Foot in Diabetic Patients. In
System Sciences (HICSS), 2013 46th Hawaii
International Conference on (pp. 2646-2655). IEEE.
Chen, J. X., Su, S. L., & Chang, C. H. (2010, July).
Diabetes care decision support system. In Industrial
and Information Systems (IIS), 2010 2nd Internationa
Conference on (Vol. 1, pp. 323-326). IEEE.
Chen, M., & Hadzic, M. (2010, October). Towards a
methodology for Lipoprotein Ontology. In Computer-
Based Medical Systems (CBMS), 2010 IEEE 23rd
International Symposium on (pp. 415-420). IEEE.
Chen, R. C., Bau, C. T., & Huang, Y. H. (2010, July).
Development of anti-diabetic drugs ontology for
guideline-based clinical drugs recommend system
using OWL and SWRL. In Fuzzy Systems (FUZZ),
2010 IEEE International Conference on (pp. 1-6).
IEEE.
Chen, R. C., Chiu, J. Y., & Bau, C. T. (2011, July). The
recommendation of medicines based on multiple
criteria decision making and domain ontology—An
example of anti-diabetic medicines. In Machine
Learning and Cybernetics (ICMLC), 2011
International Conference on (Vol. 1, pp. 27-32). IEEE.
AnIntelligentInferenceEngineusingOntologybasedClinicalPathwaysforDiagnosisandManagementofDiabetes
149

Hajja, A., Wieczorkowska, A. A., Ras, Z. W., &
Gubrynowicz, R. (2013). Pair-Based object-driven
action rules. In New Frontiers in Mining Complex
Patterns (pp. 79-93). Springer Berlin Heidelberg.
Islam, S., Freytag, G., & Shankar, R. (2012, August).
Intelligent health information system to empower
patients with chronic diseases. In Information Reuse
and Integration (IRI), 2012 IEEE 13th International
Conference on (pp. 734-737). IEEE.
Kurozumi, K., Lan, S. T., Wang, M. H., Lee, C. S.,
Kawaguchi, M., Tsumoto, S., & Tsuji, H. (2013, July).
FML-based Japanese diet assessment system. In Fuzzy
Systems (FUZZ), 2013 IEEE International Conference
on (pp. 1-6). IEEE.
Lee, C. S., Wang, M. H., & Hagras, H. (2010). A type-2
fuzzy ontology and its application to personal diabetic-
diet recommendation. In Fuzzy Systems, IEEE
Transactions on, 18(2), 374-395.
Lin, Y. (2011). Refining Ontology for Glucose
Metabolism Disorders. ICBO.
McGarry, K., Garfield, S., & Wermter, S. (2007, June).
Auto-extraction, representation and integration of a
diabetes ontology using Bayesian networks. In
Computer-Based Medical Systems, 2007. CBMS'07.
Twentieth IEEE International Symposium on (pp. 612-
617). IEEE.
Nimmagadda, S. L., Nimmagadda, S. K., & Dreher, H.
(2011, July). Multidimensional data warehousing
&mining of diabetes & food-domain ontologies for e-
Health. In Industrial Informatics (INDIN), 2011 9th
IEEE International Conference on (pp. 682-687).
IEEE.
HEALTHINF2014-InternationalConferenceonHealthInformatics
150
