
Using Ontology-based Registry and SPARQL Engine in Searching
Patient’s Clinical Documents
Juha Puustjärvi
1
and Leena Puustjärvi
2
1
Department of Computer Science, University of Helsinki, P.O. Box 68, Helsinki, Finland
2
The Pharmacy of Kaivopuisto, Neitsytpolku 10, Helsinki, Finland
Keywords: IHE XDS, SPARQL, Hl7 CDA, Electronic Health Record, Ontologies.
Abstract: As a patient may live in many places and use many healthcare specialities, patient’s clinical documents are
often stored in several systems and locations. In order to alleviate this problem, an industry initiative IHE
XDS allows health care documents to be shared over a wide area network, between hospitals, primary care
providers, and social services. Its main innovation is the logical and physically separation of the indexing
information used to retrieve documents from the actual content. Technically the XDS document registry is a
subset of the ebXML Registry standard, and documents are exchanged using SOAP and HTTP, while SQL
is used for information retrieval. Although IHE XDS has proven to be useful and workable innovation, we
have investigated whether the technologies behind the IHE XDS could be replaced by new technologies
such as by OWL-based registries and SPARQL engines. It turned out that these technologies enable the
introducing simpler policies in document exchanges. For example, contrary to the IHE XDS, we do not
have to expect patients’ records to follow then when they move from one affinity domain to another. Instead
one SPARQL query processed by a SPARQL engine is able to composing the links to patient’s original
clinical documents.
1 INTRODUCTION
An electronic health record (EHR) describes the
systematic documentation of a single patient's
medical history and care across time within one
particular health care provider's jurisdiction (Hartley
and Jones, 2005). It includes a variety of types of
observations entered over time by health care
professionals, recording observations and
administrations of drugs and therapies, orders for the
administration of drugs and therapies, test results, x-
rays, and reports.
There are many standards, such as HL7 CDA
(HL7, 2004), EN 13606 (prEN13606, 2006) and
openEHR (openEHR, 2013) developed to digitally
represent clinical data. These standards aim to
structure and markup the clinical content for the
purpose of exchange (NEHTA, 2006).
A well-known problem is that patient’s EHRs are
often stored in several systems (Puustjärvi and
Puustjärvi, 2009). This is a consequence of living in
various places, and having many healthcare
providers, including primary care physician,
specialist, therapists and other medical practitioners.
However, although patient’s clinical documentation
is stored in several EHR systems all relevant
documents should be easily accessible for the
physicians treating the patient.
The problem of patients’ scattered clinical
documents is studied in the context of Personal
Health Records (PHRs) (Raisinghani and Young,
2008), EHR archives (Hartley and Jones, 2005) and
IHE XDS (IHE, 2005). With PHRs and EHR copies
of patient´s health documentation is collected
together in advance while in IHE XDS original
documents are dynamically retrieved by exploiting
relevant registries.
In IHE XDS terminology healthcare enterprises
that agree to work together for clinical document
sharing is called clinical affinity domain. Its
enterprises agree on a common set of policies such
as how the patients are identified, the access is
controlled, and the common set of coding terms to
represent the metadata of the documents. Further,
patients expect their records to follow them as they
move from one clinical affinity domain to another.
Examples of XDS clinical affinity domains
include: nationwide and regional EHRs, federations
151
Puustjärvi J. and Puustjärvi L..
Using Ontology-based Registry and SPARQL Engine in Searching Patient’s Clinical Documents.
DOI: 10.5220/0004716501510158
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2014), pages 151-158
ISBN: 978-989-758-010-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

of enterprises, regional federations made up of
several local hospitals, healthcare providers, and
insurance provider supported communities
The key point in IHE XDS is the logical and
physically separation of the indexing information
used to retrieve documents from the actual content.
The document registry indexes documents, support
document search, and maintains a URI link back
where the document is stored in a document
repository. The basic XDS has been refined to
support special requirements for DICOM images,
structured laboratory reports, and HL7 CDA medical
summaries (CCD, 2009). The format of the used
metadata is largely based on HL7 Version 2 (Dolin
et al., 2001). Technically the XDS document registry
is a subset of the ebXML Registry standard
(ebXML, 2012), and documents are exchanged
using SOAP and HTTP (Singh and Huhns, 2005),
while SQL (Ullman and Widom, 1997) is used for
information retrieval in registries.
Although the IHE XDS has proven to be useful
and workable innovation, we argue that by
exploiting modern information technology we can
avoid many of the drawbacks of the IHE XDS. In
particular, we have addressed the following two
problems of the IHE XDS.
The main problem with ebXML registries is that
searches can only be based on the keywords and
folders. Although the keywords are taken from a
taxonomy only a very limited amount of semantics
can be provided (Dogac et al., 2007). Folders group
the related documents together (e.g., based on a
period of time, episode, or immunizations).
However, there are numerous cases where retrieving
predefined folders are not appropriate but rather
dynamic grouping of documents should be possible.
Another problem with the IHE XDS is that it
expects patients’ records to follow then when they
move from one affinity domain to another. The
problem here are twofold: First, moving records
between affinity domains is technically complicated
and error-prone due to the heterogeneities of affinity
domains. Second, due to the failed or missed
transmissions patients’ EHRs are incomplete.
We have designated a registry mechanism for
clinical documents that eliminates these drawbacks.
The expression power of document retrieval is
increased by introducing a specific OWL-ontology
(OWL, 2011), called Registry Ontology, for
document retrieval. It is derived from the class
diagram on which the Header of the HL7 CDA
documents (Boone, 2011) is based on. The primary
purpose of the CDA Header is to provide
unambiguous, structured metadata about the
document itself, which can be used in document
registers to classify, find and retrieve documents.
In our solution, the exchange of documents
between clinical affinity domains is eliminated by
retrieving all clinical documents from their original
sources. Such a feature can be carried out by the
Registry ontology and the Federated Queries
supported by SPARQL engines (SPARQL, 2008). A
useful feature of Federated Queries is that within a
query many registries can be accessed.
The rest of the paper is organized as follows.
First, in Section 2, we consider the basic
components of the IHE XDS architecture. In Section
3, we present the way the Registry Ontology is
derived. First, we give an overview of the HL7 RIM,
and the ways the RMIMs (Refined Message
Information Models) are derived from the RIM.
Then, we present the RMIM on which the Header of
the CDA documents is based on, and transform it
into OWL ontology (i.e., to Registry Ontology).
Further, in Section 4, based on the ontology we
present a SPARQL query which retrieves the URLs
of patients’ documents that are stored in two
repositories. Section 5 concludes the paper by
discussing our future work.
2 IHE XDS
2.1 IHE XDS Architecture
Integrating Healthcare Enterprise (IHE) was
established in 1999 by the Healthcare Information
Systems and Management Society (HIMSS) and the
radiological Society of North America (RSNA) to
improve the way healthcare computer systems share
information (IHE, 2005). It is not a standards
organization. Instead it promotes coordinated use of
existing standards to develop workflow solutions for
the healthcare enterprises. IHEs starting point was
radiology, where it developed profiles which specify
how to use DICOM and HL7 together, and later on it
has moved to cardiology, clinical laboratories, and
other specialities.
Another dimension of IHE’s work has been the
development of IT infrastructures standards for use
across departmental and institutional boundaries.
The IHE XDS profile is an example of this. Systems
designed in agreement with IHE profiles
communicate better with one another, and facilitate
efficient access to information.
The idea behind the IHE XDS is to build virtual
patient records on the fly from a variety of clinical
documents created by different healthcare
HEALTHINF2014-InternationalConferenceonHealthInformatics
152
organizations (Benson, 2010). The separation of the
metadata (indexing information) used to retrieve
documents from the actual content allows IHE XDS
to handle any type of content and simplifies the
addition of an XDS export function to existing
systems. Thereby each document is viewed in its
original form.
The components of the IHE XDS are document
source, document repository, document registry and
the document consumer. The IHE XDS architecture
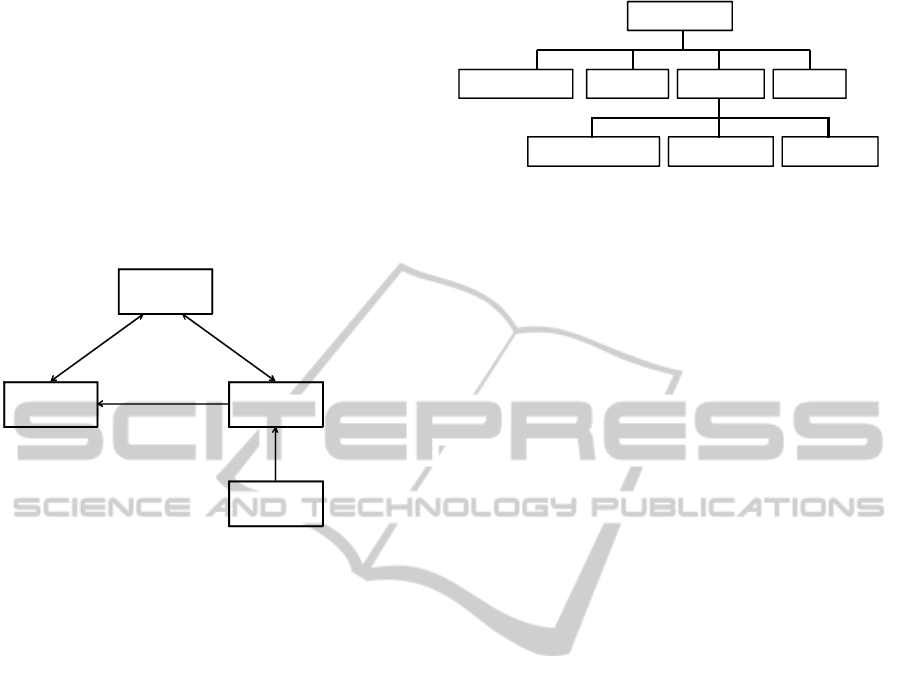
within one affinity domain is presented in Figure 1.
Document
Consumer
Document
Registry
Document
Repository(ies)
Document
Source(s)
Query Documents
Retrieve Documents
Register Document
Provide and
Register Document Set
Figure 1: IHE XDS architecture.
The document source produces original documents,
submits these to a document repository, and also
produces metadata about each stored document
which is sent to the document registry. There may be
one or more document repositories and each
provides secure document storage and supports
document retrieval. Documents may be organized in
folders.
The document registry indexes documents,
support document search, and maintains a URI link
back where the document is stored in a document
repository. The document consumer is a user system.
It initiates searches of the register, retrieves and
displays selected documents from their repositories.
2.2 ebXML Registry Information
Model
The ebXML Registry Information Model defines
what types of objects are stored in the registry and
how they are organized (Dogac et al., 2007). Figure
2 represents the hierarchical structure of the ebXML
registry constructs.
Top level class “RegistryObject” provides
minimal metadata for registry objects. Other
instances are used to provide a dynamic way to add
arbitrary attributes to “RegistryObject” instances.
For example, Association instances can be used to
define many-to-many associations between objects
RegistryObject
ClassificationNode Classification RegistryEntry Association
ClassificationScheme RegistryPackage ExtrinsicObject
Figure 2: Components of the ebXML Registry Information
Model.
in the information model. Further, each association
has an “associationType” attribute that identifies the
type of that association.
There are also many predefined Association
Types and new types can be introduced when
needed. As a result the ebXML RIM structures
enable the specification of conceptual schemas.
Further “ClassificationScheme” instances describe a
structured way to classify and categorize
RegistryObject instances. RegistryPackage instances
group logically related RegistryObject instances
together, and thus enable the specification of the
folders of the clinical documents.
2.3 Enhancing IHE XDS
To facilitate information exchange between
heterogeneous clinical affinity domains the IHE
XDS is enhanced in (Dogac et al., 2002) by a
specific ontology which enables the mappings
between heterogeneous affinity domains. The
ontology is first specified in OWL, and then
transformed in the format of ebXML Registry
Information Model, and finally it is transformed into
relations and stored in the ebXML registries that can
be queried by SQL.
Thus the document discovery across affinity
domains is facilitated through ontology mappings
when affinity domain specific metadata is defined
through the ontology (Dogac et al., 2007). The
introducing of the ontology enables the specification
of the associations between the nodes. The
associations are defined by modeling primitives
supported by the ebXML Registry Information
Model, and are stored in the Registry.
UsingOntology-basedRegistryandSPARQLEngineinSearchingPatient'sClinicalDocuments
153

3 HL7 CLINICAL DOCUMENT
ARCHITECTURE
3.1 HL7 Reference Information Model
The HL7 Reference Information Model (RIM) is the
cornerstone of the HL7 message development
process and development methodology (Puustjärvi
and Puustjärvi, 2010). It expresses the data content
needed in a specific clinical or administrative
context and provides an explicit representation of the
semantic and syntactical connections that exist
between the information carried in the fields of HL7
messages (Boone, 2011).
The RIM is based on two key ideas (Benson,
2010). The first idea is based on the consideration
that most healthcare documentation is concerned
with “happenings” and things (human or other) that
participate in these happenings in various ways.
The second idea is the observation that the same
people or things can perform different roles when
participating in different types of happening, e.g., a
person may be a care provider such a physician or
the subject of care such as patient.
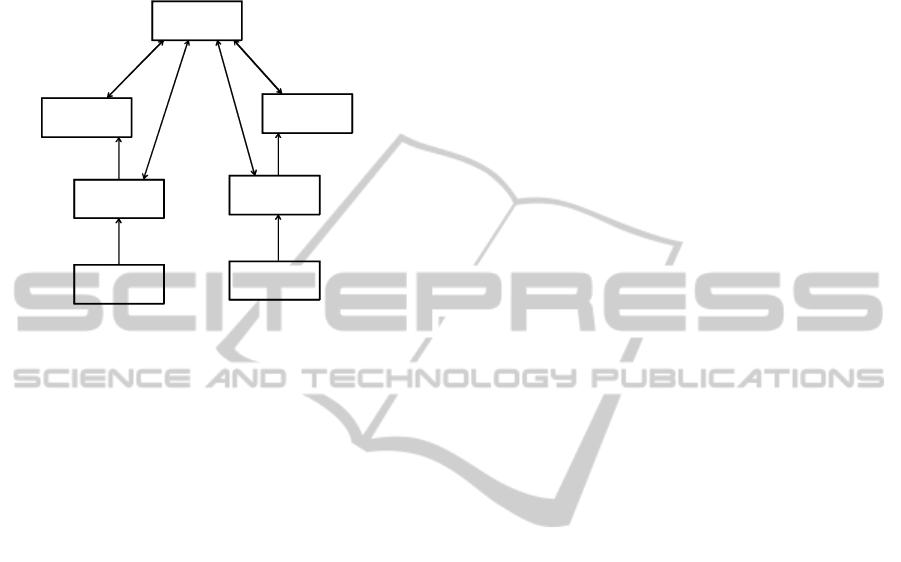
As a result of these ideas the RIM is based on a
simple backbone structure, involving three main
classes, Act, Role, and Entity, linked together using
three association classes Act-Relationship,
Participation, and Role-Relationship (Figure 3).
Note that HL7 uses its own representation of UML
to reflect the use of these six backbone classes. Each
class has its own color and shape to represent the
stereotypes of these classes, and they only connect in
certain ways.
Figure 3: RIM backbone structure.
Each happening is an Act and it may have any
number of Participations, which are Roles, played by
Entities. An ACT may also be related to other Acts
via Act Relationships. Act, Role and Entity classes
have a number of specializations (subclasses), e.g.,
Entity has a specialization LivingSubject, which
itself has a specialization Person.
The classes in the RIM have structured attributes
which specify what each RIM class means when
used in a message (exchanged document). For
example, Act has structured attributes classCode and
moodCode. The former states what sort of Act this is
(e.g., observation, encounter, or administration of a
drug). moodCode indicates whether an Act has
happened, is request for something to happen, a goal
or a criterion. The idea behind structured attributes is
to reduce the original RIM from over 100 classes to
a simple backbone of six main classes (Benson,
2010).
3.2 Refined Message Information
Model RMIM
The RIM is not a model of healthcare, nor is it a
model of any message, although it is used in
exchanged messages. The structures of exchanged
documents are defined by constrained information
models (HL7, 2007). The most commonly used
constrained information model is the RMIM. Each
RMIM is a diagram that specifies the structure of an
exchanged document.
A RMIM diagram is specified for a specific use
case (Boone, 2011). The diagram is derived from the
RIM by limiting its optionality. Such specifications
are called CDA Profiles (Spronk, 2008).
In developing a RMIM diagram the RIM is
constrained by omission and cloning. Omission
means that the RIM classes or attributes can be left
out. Note that all classes and attributed that are not
structural attributes in the RIM are optional, and so
the designer can take only the needed classes and
attributes. Cloning means that the same RIM class
can be used many times in different ways in various
RMIMs. The classes selected for a RMIM are called
clones.
The multiplicities of associations and attributes
in a RMIM are constrained in terms of repeatability
and optionality. Further, code binding is used for
specifying the allowable values of the used
attributes.
Although the semantics of all CDA documents is
tractable through a RMIM back to the RIM, we
neither can use the RMIM nor the RIM in
formulating queries on patient’s health
documentation as each RMIM only models one type
of documents. Another reason is that there are no
query languages specified for the information model
used in the RMIM and RIM schemas.
3.3 CDA Levels
Each Continuity of Care document (CCD) has one
primary purpose (which is the reason for the
generation of the document), such as patient
HEALTHINF2014-InternationalConferenceonHealthInformatics
154

admission, transfer, or inpatient discharge. Each
Clinical Document Architecture (CDA) document is
made up of the header and the body (Benson, 2010).
Depending whether the header and body of the
CDA documents are based on the RIM they are
classified into three levels:
CDA Level 1: Only the header is based on the RIM
while the body is human readable text or image.
CDA Level 2: Only the header of the document is
based on the RIM while the body is comprised of
XML coded sections.
CDA Level 3: Both the header and the body are
based on the RIM.
The CDA header is common to all the three
levels of CDA. The header contains basic metadata.
These include information about what the
documents is, who created it, when, where, and for
what purposes. Its primary purpose is to provide
unambiguous, structured metadata about the
document itself, which can be used in document
registers to classify, find and retrieve documents.
In HL7 CDA terminology the header is an
instance of an Act called Clinical Document. This
means that there is a Refined Message Information
Model (RMIM) that models the headers of all HL7
CDA documents. To illustrate this, a simplified
RMIM of the Header of CDA documents is
presented in Figure 4. The diagram presents classes
of the RMIM but not all their attributes.
ClinicalDocument
Subject Performer
Patient
Employee
Person Organization
1..1 patientPerson 1..1employeeOrganization
CDAHeader
classCode
moodCode
Id
code
classCode
name
classCode
Id
classCode
Id
classCode
Id
Figure 4: A simplified RMIM of CDA Header.
Note that HL7 uses its own representation of UML
in RMIM diagrams: each class has its own colour
and shape to represent the stereotypes of these
classes, and they only connect in certain ways.
The entry point of this diagram (CDA Header) is
ClinicalDocument, which is specialization of the
RIM class Act. Classes Patient and Employee are
specializations (subclasses) of the RIM class Role.
Person and Organization are specializations of the
RIM class Entity. Subject and Performer are
specializations of the association class Participation.
Each specialization inherits all of the properties
(attributes) of the generalization. For example, the
class Patient is a specialization of Role with the
addition of the optional attribute
veryImportantPersonCode.
3.4 Transforming HL7 CDA Header
into OWL
Although the semantics of all CDA documents is
traceable through a RMIM back to the RIM, we
neither can use a RMIM nor the RIM in formulating
queries as there are no query languages specified for
the information model used in the RMIM and RIM
schemas. For this reason we transform the RMIM of
the CDA header into Web Ontology Language
(OWL) (OWL, 2011).
Transforming a RMIM diagram into OWL is
straightforward in the sense that both models are
object-oriented although the notation used in RMIM
diagrams slightly differs from the traditional UML
notation. Yet their basic modelling primitives are the
same, namely classes, subclasses, properties and
values. The classes are also connected in a similar
way through properties.
In order to illustrate the transformation of RMIM
diagram into OWL we have presented the RMIM
diagram of Figure 4 in OWL in Figure 5.
Classes, subclasses, data properties and object
properties are modeling primitives in OWL
(Antoniou and Harmelen, 2004). Object properties
relate objects to other objects while datatype
properties relate objects to datatype values (Daconta
et al, 2003). For example, Performer is an object
property. Its domain is clinicalDocument and range
is employee. Note that, in Figure 5 we have omitted
most datatype properties. The only datatype property
presented in the figure is “code”. Its domain is
clinicalDocumernt and its range is xsd:string, i.e.,
string in “XML-terminology”.
UsingOntology-basedRegistryandSPARQLEngineinSearchingPatient'sClinicalDocuments
155

<rdf:RDF
xmln s:rdf=http://www.w3.org/1999/02/22-rdf-syntax-nsl#
xmln s:rdfs=http://www.w3.org/2000/01/rdf-schema#
xmln s:owl=http://www.w3.org/2002/07/owl#>
xmln s:xsd =http://www.w3.org/2001/xml-schemal#>
<owl:On tology rd f:about=“registryOntology”/>
<owl:Class rdf:ID=“act/”>
<owl:Class rdf:ID=“role/”>
<owl:Class rdf:ID=“entity/”>
<owl:Class rdf:ID=“participation/”>
<owl:Class rdf:ID=“clinicalDocument”>
<rdfs: subClassOf rdf:: resource “#act”/>
</owl :class>
<owl:Class rdf:ID=“patient”>
<rdfs: subClassOf rdf:: resource “#role”/>
</owl :class>
<owl:Class rdf:ID=“employee”>
<rdfs: subClassOf rdf:: resource “#role”/>
</owl :class>
<owl:Class rdf:ID=“person”>
<rdfs: subClassOf rdf:: resource “#entity”/>
</owl :class>
<owl:Class rdf:ID=“organization”>
<rdfs: subClassOf rdf:: resource “#entity”/>
</owl :class>
<owl:ObjectProperty rdf:ID=“subject”>
<rdfs:d omain rd f:resource=“#clinicalDocument”/>
<rdfs:ran ge rd f:resource=“#patient”/>
</owl:ObjectProperty>
<owl:ObjectProperty rdf:ID=“patientPerson”>
<rdfs:domain rd f:resource=“#patient”/>
<rdfs:ran ge rd f:resource=“#person”/>
</owl:ObjectProperty>
<owl:ObjectProperty rdf:ID=“performer”>
<rdfs:d omain rd f:resource=“#clinicalDocument”/>
<rdfs:rangerdf:resource=“#employee”/>
</owl:ObjectProperty>
<owl:ObjectProperty rdf:ID=“employeeOrganization”>
<rdfs:domain rd f:resource=“#employee”/>
<rdfs:ran ge rd f:resource=“#organization”/>
</owl:ObjectProperty>
<owl:Datatyp eProperty rdf:ID=“code”>
<rdfs:domain rd f:resource=“#lclinicalDocument”/>
<rdfs:ran ge rd f:resource=“&xsd;string”/>
</owl:Datatyp eProperty>
.
.
.
</rdf:RDF>
Figure 5: A part of CDA Header in OWL.
4 QUERYING CLINICAL
DOCUMENTS BY SPARQL
4.1 SPARQL Queries
The name SPARQL is a recursive acronym for
SPARQL Protocol and RDF Query Language, which
is described by a set of specifications from the W3C
(DuCharme, 2011). SPARQL Protocol refers to the
rules for how a client program and a SPARQL
processing server exchange SPARQL queries and
results.
A typical SPARQL query specifies the pieces of
information that meets the stated conditions. The
conditions are described with triple patterns, which
are similar to RDF triples but may include variables
to add flexibility in how they match against the data.
There is a variety of SPARQL processors (also
called SPARQL engines) available for running
queries against data both locally and remotely.
SPARQL provides two ways for querying remotely:
using FROM keyword or using SERVICE keyword.
In the former way the FROM keyword names a
dataset to query that may be local or remote file. In
the latter way, instead of pointing at an RDF file
somewhere, a SPRQL endpoint is pointed. A
SPARQL endpoint is a web service that accepts
SPARQL queries, runs the queries, and then returns
the result.
4.2 Federated Queries
Federated Queries in SPARQL allow searching
multiple datasets with one query. For each dataset is
created a subquery which access datasets by using
SERVICE keywords. That is, federated SPARQL
queries make use of subqueries and SERVICE
keywords. To illustrate this consider the federated
SPARQL query presented in Figure 6. The query is
based on the ontology presented in Figure 5, and the
prefix ro in the query refers to that ontology. The
query returns the addresses of Lisa Smith’s clinical
documents by accessing two datasets through
SPARQL endpoints. The result of the query is the
union of the results of the two subqueries.
PREFIXowl:<http://www.w3.org/2002/07/owl#>
PREFIXrdf:<http://www.w 3.org/1999/02/22‐ rdf‐syntax‐ ns#>
PREFIXrdfs:<http://w w w.w3.org/2000/01/rdf‐ schema#>
PREFIXro:<http://w w w.cs.helsinki.fi/registry Ontology#>
PREFIXpe :<http://w w w.healthstore/resource/people/>
SELECT?documentAddress
WHERE
{
SERVICE<http://documentRegistry_A/sparql>
{SELECT?id
WHERE
{
pe:Lisa_ Smith ro:subject ?id;
?idro:classCode ro:O bservation ;
?idro:code “71620000”.
}
}
SERVICE<http://documentRegistry_B/sparql>
{SELECT?documentAddress
WHERE
{
pe:Lisa_ S m ith ro :subject ?id;
?idro:classCode ro:O bservation ;
?idro:code “71620000”.
}
}
}
Figure 6: A simple federated SPARQL query.
This SPARQL query is based on the architecture
presented in Figure 7. The query of Figure 6
HEALTHINF2014-InternationalConferenceonHealthInformatics
156
presents the communication between the document
consumer and the two document registries (denoted
by “Query Documents” in the figure). Documents
Registries provide SPARQL endpoints, which are
web services.
Document
Consumer
Document
Registry
Document
Repository
Document
Source(s)
Query Documents
Retrieve
Documents
Register Document
Provide and
Store Document s
Query Documents
Document
Registry
Document
Repository
Document
Source(s)
Register Document
Provide and
Store Document s
...
Figure 7: The Architecture of the communicating
components.
Note that in our solution all document registries are
accessed in querying the locations of patient’s
documents. As a result, many of the registries are
queried though they have no registry items of the
patient. Such unnecessary queries could be avoided
if the Document Consumer is informed about the
relevant registries or a dictionary is maintained
which includes such information.
5 CONCLUSIONS
Patient’s clinical documents are often stored in
several healthcare providers’ systems. This is a
consequence of living in various places, and having
many healthcare providers, including primary care
physician, specialist, therapists and other medical
practitioners.
The problem of patients’ scattered clinical
documentation can be managed by using personal
health records, EHR archives and IHE XDS
Registries.
The key point in IHE XDS is the logical and
physically separation of the indexing information
used to retrieve documents from the actual content.
The document registry indexes documents, support
document search, and maintains a URI link back
where the document is stored in a document
repository. Further, the Cross-Community Patient
Discovery (XCPD) profile supports the means to
locate communities which hold patient relevant
health data and the translation of patient identifiers
across communities holding the same patient’s data.
Although the IHE XDS has proven to be useful
innovation, we argue that its used ebXML Registry
standard does not provide enough semantics for
indexing clinical documents. Instead, according to
ebXML original purpose, its data structures are
appropriate for indexing and classifying the web
services of electronic business.
Our studies have shown that building a clinical
document registry based on the class diagram of the
HL CDA Header is a logical choice as its original
purpose is to provide metadata for document
registries. Further, SPARQL language and
SPARQL engines provide an elegant way for
accessing several registries within a query. As a
result, we do not have to expect that patients’
records follow them as they move from one clinical
domain to another.
For now, our proposed solution is restricted on
CDA documents as they already provide sufficient
metadata for the registries. However, our solution
can be easily extended to other formats, such as
DICOM and PDF, by annotating these documents in
a way that is compliant with the CDA document
header.
In our future work, we will investigate whether it
is possible to extend the semantics of the Registry
Ontology, which is now based on the CDA Header.
The problem here is that the efficient usage of
patients’ health documentation often is data centric,
meaning that documents should be retrieved based
on their content. For example, a physician may be
interested to retrieve the documents dealing with
Emconcor (a drug for blood pressure). However, the
data required by such queries is not provided by the
CDA Header but rather it is provided by the CDA
Body. This suggests that it may be reasonable to
extend the Registry Ontology by some elements
taken from the CDA Body.
REFERENCES
Antoniou, G., Harmelen, F., 2004. A Semantic Web
Primer. The Mitt Press.
Benson, T., 2010. Principles of Health Interoperability
HL7 and SNOMED. Springer.
Boone, K., 2011. The CDA Book. Springer.
CCD, 2009. What Is the HL7 Continuity of Care
Document? Available at: http://www.neotool.com/
blog/2007/02/15/ what-is-hl7 -continuity -of- care-
document/
UsingOntology-basedRegistryandSPARQLEngineinSearchingPatient'sClinicalDocuments
157

Daconta, M., Obrst, L., Smith, K., 2003. The semantic
web: A Guide to the Future of XML, Web Services,
and Knowledge Management, John Wiley & Sons.
Dogac, A., Laleci, G., Kabak, Y., Cingil, I., 2002,
Exploiting web service semantics: Taxonomies vs
ontologies, IEEE Data Eng. Bull, Vol. 25, No. 4,
pp.10-16.
Dogac, A., Gokce, B., Aden, T., Laleci, T., Eichelberg,
M. 2007. Enhancing IHE XDS for Federated Clinical
Affinity Domain Support. IEEE Transactions on
Information Technology in Biomedicine, Vol.11, No.
2.
Dolin, R., Alschuler, L., Beerb, C., Biron, P., Boyer, S.,
Essin, E., Kimber, T. 2001. The HL7 Clinical
Document Architecture. J. Am Med Inform Assoc,
2001:8(6), pp. 552-569.
DuCharme, 2011. Learning SPARQL. O’Reilly Media.
ebXML, 2012, ebXML Registry Standards, 2012.
Available at: http://ebxml.xml.org/standards.
Hartley, C., Jones, E., 2005. EHR Implementation. AMA
Press.
HL7, 2004. EHR System Functional Model and Standard
Draft Standard for Trial Use (DSTU).
HL7, 2007. What is the HL7 Continuity of Care
Document? Available at: http://www.neotool.com/
blog/2007/02/15/what-is-hl7- continuity- of- care-
document/
IHE, 2005. IT Infrastructure Technical Framework
Volume 1 (ITI TF-1). Available at:
www.ihe.net/Technical_Framework/upload/ihe_iti_tf_2.0
_vol1_FT_2005-08-15.pdf.
NEHTA. 2006. Review of shared electronic health record
standards. Version 1.0. National e- Health Transition
Authority, Available at:
.nehta.gov.au/component/option,com_docman/task,cat_vie
w http://www /gid,130/Itemid,139/
openEHR Foundation. Available at: http:
//www.openehr.org.
OWL, 2011. WEB OntologyLanguage. Available at:
http://www.w3.org/TR/owl-features/
prEN13606, 2006.Health informatics – Electronic
healthcare record communication – Parts 1-5.
Committee European Normalisation, CEN/TC 251
Health Informatics Technical Committee. Available
at: http://www.centc251.org/
Puustjärvi, J., Puustjärvi, L., 2009. The role of medicinal
ontologies in querying and exchanging pharmaceutical
information. International Journal of Electronic
Healthcare, Vol. 5, No.1 pp. 1 – 13.
Puustjärvi, J., Puustjärvi, L., 2010. Automating the
Importation of Medication Data into Personal Health
Records. In the proc. of the International Conference
on Health Informatics. Pages 135-141.
Raisinghani M.S., Young, E., 2008. Personal health
records: key adoption issues and implications for
management, International Journal of Electronic
Healthcare. Vol. 4, No.1 pp.67-77.
Singh., M., Huhns, M., 2005. Service Oriented
Computing. Wiley.
SPARQL, 2008. SPARQL Query Language for RDF.
Available at: http://www.w3.org/TR/rdf-sparql-query/
Spronk, R.. 2008. CDA around the world: archive of CDA
instance examples, Availble at:
http://www.ringholm.com/download/CDA_R2_examp
les.zip.
Ullman, D., Widom, J., 1997. A First Course in Database
Systems. Prentice Hall.
HEALTHINF2014-InternationalConferenceonHealthInformatics
158
