
Unsupervised Consensus Functions Applied to Ensemble Biclustering
Blaise Hanczar and Mohamed Nadif
LIPADE, University Paris Descartes, 45 rue des saint-peres, 75006 Paris, France
Keywords:
Biclustering, Ensemble Methods, Consensus Functions.
Abstract:
The ensemble methods are very popular and can improve significantly the performance of classification and
clustering algorithms. Their principle is to generate a set of different models, then aggregate them into only
one. Recent works have shown that this approach can also be useful in biclustering problems.The crucial
step of this approach is the consensus functions that compute the aggregation of the biclusters. We identify
the main consensus functions commonly used in the clustering ensemble and show how to extend them in
the biclustering context. We evaluate and analyze the performances of these consensus functions on several
experiments based on both artificial and real data.
1 INTRODUCTION
Biclustering, also called direct clustering (Hartigan,
1972), simultaneous clustering in (Govaert, 1995;
Turner et al., 2005) or block clustering in (Govaert
and Nadif, 2003) is now a widely used method of data
mining in various domains in particular in text mining
and bioinformatics. For instance, in document clus-
tering, in (Dhillon, 2001) the author proposed a spec-
tral block clustering method which makes use of the
clear duality between rows (documents) and columns
(words). In the analysis of microarray data, where
data are often presented as matrices of expression lev-
els of genes under different conditions, the co- clus-
tering of genes and conditions overcomes the problem
encountered in conventional clustering methods con-
cerning the choice of similarity. Cheng and Church
(Cheng and Church, 2000) were the first to propose
a biclustering algorithm for microarray data analy-
sis. They considered that biclusters follow an addi-
tive model and used a greedy iterative search to mini-
mize the mean square residue (MSR). Their algorithm
identifies the biclusters one by one and was applied to
yeast cell cycle data, and made it possible to identify
several biologically relevant biclusters. Lazzeroni and
Owen (Lazzeroni and Owen, 2000) have proposed
the popular plaid model which has been improved by
Turner et al. (Turner et al., 2005). The authors as-
sumed that biclusters are organized in layers and fol-
low a given statistical model incorporating additive
two way ANOVA models. The search approach is it-
erative: once (K − 1) layers (biclusters) were identi-
fied, the K-th bicluster minimizing a merit function
depending on all layers is selected. Applied to data
from the yeast, the proposed algorithm reveals that
genes in biclusters share the same biological func-
tions. In (Erten and S
¨
ozdinler, 2010) the authors de-
veloped their localization procedure which improves
the performance of a greedy iterative biclustering al-
gorithm. Several other methods have been proposed
in the literature, two complete surveys of biclustering
methods can be found in (Madeira and Oliveira, 2004;
Busygin et al., 2008).
Here we propose to use the ensemble methods to
improve the performance of biclustering. It is impor-
tant to note that we do not propose a new biclustering
method in competition with the previously mentioned
algorithms. We seek to adapt the ensemble approach
to the biclustering problem in order to improve the
performance of any biclustering algorithm. The prin-
ciple of ensemble biclustering is to generate a set of
different biclustering solutions, then aggregate them
into only one solution. The crucial step is based on
the consensus functions computing the aggregation of
the different solutions. In this paper we have identi-
fied four types of consensus function commonly used
in ensemble clustering and giving the best results. We
show how to extend their use in the biclustering con-
text. We evaluate their performances on a set of both
numerical and real data experiments.
The paper is organized as follows. In Section 2,
we review the ensemble methods in clustering and bi-
clustering. In section 3, we formalize the collection
of biclustering solutions and show how to construct
it from the Cheng & Church algorithm that we chose
for our study. In section 4, we extend four commonly
30
Hanczar B. and Nadif M..
Unsupervised Consensus Functions Applied to Ensemble Biclustering.
DOI: 10.5220/0004789800300039
In Proceedings of the 3rd International Conference on Pattern Recognition Applications and Methods (ICPRAM-2014), pages 30-39
ISBN: 978-989-758-018-5
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

used consensus functions to the biclustering context.
Section 5 is devoted to evaluate these new consen-
sus functions on several experimentations. Finally,
we summarize the main points resulting from our ap-
proach.
2 ENSEMBLE METHODS
The principle of ensemble methods is to construct a
set of models, then to aggregate them into a single
model. It is well-known that these methods often per-
form better than a single model (Dietterich, 2000).
Ensemble methods first appeared in supervised learn-
ing problems. A combination of classifiers is more
accurate than single classifiers (Maclin, 1997). A pi-
oneer method boosting, the most popular algorithm
which is adaboost, was developed mainly by Shapire
(Schapire, 2003). The principle is to assign a weight
to each training example, then several classifiers are
learned iteratively and between each learning step the
weight of the examples is adjusted depending on the
classifier results. The final classifier is a weighted
vote of classifiers constructed during the procedure.
Another type of popular ensemble methods is bag-
ging, proposed by Breiman (Breiman, 1996). The
principle is to create a set a classifiers based on boot-
strap samples of the original data. The random forests
(Breiman, 2001) are the most famous application of
bagging. They are a combination of tree predictors,
and have given very good results in many domains
(Diaz-Uriarte and Alvarez de Andres, 2006).
Several works have shown that ensemble methods
can also be used in unsupervised learning. Topchy et
al. (Topchy et al., 2004b) showed theoretically that
ensemble methods may improve the clustering per-
formance. The principle of boosting was exploited
by Frossyniotis et al. (Frossyniotis et al., 2004) in
order to provide a consistent partitioning of the data.
The boost-clustering approach creates, at each itera-
tion, a new training set using weighted random sam-
pling from original data, and a simple clustering algo-
rithm is applied to provide new clusters. Dudoit and
Fridlyand (Dudoit and Fridlyand, 2003) used bagging
to improve the accuracy of clustering in reducing the
variability of the PAM algorithm (Partitioning Around
Medoids) results (van der Laan et al., 2003). Their
method has been applied to leukemia and melanoma
datasets and made it possible to differentiate the dif-
ferent subtypes of tissues. Strehl et al. (Strehl and
Ghosh, 2002) proposed an approach to combine mul-
tiple partitioning obtained from different sources into
a single one. They introduced heuristics based on a
voting consensus. Each example is assigned to one
cluster for each partition, an example has therefore as
many assignments as number of partitions in the col-
lection. In the aggregated partition, the example is
assigned to the cluster to which it was the most of-
ten assigned. One problem with this consensus is that
it requires knowledge of the cluster correspondence
between the different partitions. They also proposed
a cluster-based similarity partitioning algorithm. The
collection is used to compute a similarity matrix of
the examples. The similarity between two examples
is based on the frequency of their co-association to
the same cluster over the collection. The aggregated
partition is computed by a clustering of the exam-
ples from the similarity matrix. Fern (Fern and Brod-
ley, 2004) formalized the aggregation procedure by a
bipartite graph partitioning. The collection is repre-
sented by a bipartite graph. The examples and clus-
ters of partitions are the two sets of vertices. An edge
between an example and a cluster means that exam-
ple has been assigned to this cluster. A partition of
the graph is performed and each sub-graph represents
an aggregated cluster. Topchy (Topchy et al., 2004a)
proposed to modelize the consensus of the collection
by a multinomial mixture model. In the collection,
each example is defined by a set of labels that rep-
resents their assigned clusters in each partition. This
can be seen as a new space in which the examples are
defined, each dimension being a partition of the col-
lection. The aggregated partition is computed from a
clustering of examples in this new space. Since the
labels are discrete variables, a multinomial mixture
model is used. Each component of the model repre-
sents an aggregated cluster.
Some recent works have shown that the ensem-
ble approach can also be useful in biclustering prob-
lems (Hanczar and Nadif, 2012). DeSmet pre-
sented a method of ensemble biclustering for query-
ing gene expression compendia from experimental
lists (De Smet and Marchal, 2011). Actually the en-
semble approach is performed only one dimension of
the data (the gene dimension). Then biclusters are ex-
tracted from the gene consensus clusters. A bagging
version of biclustering algorithms has been proposed
and tested for microarray data (Hanczar and Nadif,
2010). Although this last method improves the per-
formance of biclustering, in some cases it fails and
returns empty biclusters, i.e. without examples or fea-
tures. This is because the consensus function handles
the sets of examples and features on the same dimen-
sion as in the clustering context. The consensus func-
tion must respect the structure of the biclusters. For
this reason, the consensus functions mentioned above,
can be applied to biclustering problems. In this paper
we adapt these consensus functions to the biclustering
UnsupervisedConsensusFunctionsAppliedtoEnsembleBiclustering
31

context.
3 BICLUSTERING SOLUTION
COLLECTION
The first step of ensemble biclustering is to generate
a collection of biclustering solution. Here we give
the formalization of the collection and a method to
generate it from the Cheng and Church algorithm that
we have chosen for our study.
3.1 Formalization of the Collection
Let a data matrix be = { , } where =
{e
1
,...,e
N
} is the set of N examples represented by
M-dimensional vectors and = { f
1
,..., f
M
} is the
set of M features represented by N-dimensional vec-
tors. A bicluster B is a submatrix of X defined
by a subset of examples and a subset of features:
B = {(E
B
,F
B
)|E
B
⊆ ,F
B
⊆ }. A biclustering op-
erator Φ is a function that returns a biclustering so-
lution (i.e. a set of biclusters) from a data matrix:
Φ(X) = {B
1
,...,B
K
} where K is the number of bi-
clusters. Let ϕ be the function giving for each point
of the data matrix the label of the bicluster to which
it belongs. The label is 0 for points belonging to no
bicluster.
ϕ(x
i j
) =
{
k i f e
i
∈ E
B
k
and f
j
∈ F
B
k
0 i f e
i
/∈ E
B
k
or f
j
/∈ F
B
k
∀k ∈ [1,K].
A biclustering solution can be represented by a label
matrix I
¯
giving for each point: I
i j
= ϕ(x
i j
). In the
following it will be convenient to represent this label
matrix by an label vector indexed by u defined as u =
i ∗| | +(| | − j), where |.| denotes the cardinality. J
¯
is the vector form of the matrix I
¯
: J
¯
u
= J
¯
i∗| |+(| |− j
=
ϕ(x
i j
.
Let’s the true biclustering solution of the data set
represented by Φ( )
∗
, I
¯
∗
and J
¯
∗
. An estimated bi-
clustering solution is a biclustering solution returned
by an algorithm from the data matrix, it is denoted by
ˆ
Φ( ),
ˆ
I
¯
and
ˆ
J
¯
. The objective of the biclustering task is
to find the closest estimated biclustering solution from
the true biclustering solution. In ensemble methods,
we do not use only one estimated biclustering solu-
tions but we generate a collection of several solutions.
We denote this collection of biclustering solutions
as follows = {
ˆ
Φ(X)
(1)
,...,
ˆ
Φ(X)
(R)
}. This collec-
tion can be represented by an NM × R matrix =
(J
¯
T
1
,...,J
¯
T
NM
)
T
by merging together all label vectors
J
¯
u
= (J
u1
,...,J
uR
)
T
where J
ur
= ϕ(x
i j
)
(r)
with r ∈
[1,R]. The objective of the consensus function is to
form an aggregated biclustering solution, represented
by Φ(X), I
¯
and J
¯
, from the collection of estimated so-
lutions. Each of these functions is illustrated with an
example in Figure 1.
3.2 Construction of the Collection
The key point of the generation of the collection is
to find a good trade-off between the quality and di-
versity of the biclustering solutions of the collection.
If all the generated solutions are the same, the aggre-
gated solution is identical to the biclusters of the col-
lection. Different sources of the diversity are possible.
We can use a resampling method such as bootstrap
or jacknife. In applying the biclustering operator to
each resampled data, different solutions are produced.
We can also include the source of diversity directly in
the biclustering operator. In this case the algorithm is
not deterministic and will produce different solutions
from the same original data.
In our experiments the biclustering operator is the
Cheng and Church algorithm (CC) (algorithm 4 in the
reference (Cheng and Church, 2000)). This algorithm
returns a set of biclusters minimizing the mean square
residue ( MSR).
MSR(B
k
) =
1
|B
k
|
∑
i, j
z
ik
w
jk
(X
i j
− µ
ik
− µ
jk
+ µ
k
)
2
,
where µ
k
is the average of B
k
, µ
ik
and µ
jk
are respec-
tively the means of E
i
and F
j
belonging to bicluster
B
k
. z and w are the indicator functions of the exam-
ples and features. z
ik
= 1 when the feature i belongs
to the bicluster k, z
ik
= 0 otherwise. w
jk
= 1 when the
example j belongs to the bicluster k, w
jk
= 0 other-
wise.
The CC algorithm is iterative and the biclusters
are identified one by one. To detect each bicluster, the
algorithm begins with all the features and examples,
then it drops the feature or example minimizing the
mean square residue (MSR) of the remaining matrix.
This procedure is totally deterministic. We modified
the CC algorithm by including a source of diversity in
the computation of the bicluster. At each iteration, we
selected the top α% of the features and examples min-
imizing MSR of the remaining matrix. The element to
be dropped was randomly chosen from this selection.
Thus the parameter α controls the level of diversity
of the bicluster collection; in our simulations α = 5%
seemed a good threshold. This modified version of
the algorithm was used in all our experiments in or-
der to generate the collection of biclustering solutions
from a dataset.
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
32

4 CONSENSUS FUNCTIONS FOR
BICLUSTERING
The second step of the ensemble approach is the ag-
gregation of the collection of biclustering solutions.
We present here the extension of four consensus func-
tions for biclustering ensemble. These methods as-
sign a bicluster label to the N × M points of the data
matrix. Note that even when the numbers of biclus-
ters in the different solutions of the collection are not
equal, these consensus functions can be used; it suf-
fices to fix the final number of aggregated biclusters
to K.
4.1 CO-Association Consensus (COAS)
The idea of COAS is to group in a bicluster the points
that are assigned together in the biclustering collec-
tion. This consensus is based on the bicluster assig-
nation similarity between the points of the data ma-
trix. The similarity between two points is defined by
the proportion of times that they are associated to the
same bicluster over the whole collection. All these
similarities are represented by a distance matrix D de-
fined by:
D
uv
= 1 −
1
R
R
∑
r=1
δ(J
ur
= J
vr
),
where δ(x) returns 0 when x is false and 1 when true.
From this dissimilarity data matrix, K + 1 clusters are
identified in using the Partitioning Around Medoids
(PAM) algorithm (Dudoit and Fridlyand, 2003). The
K clusters of points represent the K aggregated biclus-
ters, the last cluster groups all the points that belongs
to no bicluster.
4.2 Voting Consensus (VOTE)
This consensus function is based on the majority vote
of the labels. Each point is assigned to the bicluster
with which it has been assigned the most of the time in
the biclustering collection. For each point of the data
matrix, the consensus returns the most represented la-
bel in the collection of the biclustering solution. The
main problem of this approach is that there is no cor-
respondence between the labels of two different esti-
mated biclustering solutions. All the biclusters of the
collection have to be re-labeled according to their best
agreement with some chosen reference solution. Any
estimated solution can be used as reference, here we
used the first one
ˆ
Φ(X)
(1)
. The agreement problem
can be solved in polynomial time by the Hungarian
method (Papadimitriou and Steiglitz, 1982) which re-
labels the estimated solution such the similarity be-
tween the solutions is maximized. The similarity be-
tween two biclustering solutions was computed by us-
ing the F-measure (details in section 5.1). The label
of the aggregated biclustering solution for a point is
therefore defined by:
J
¯
u
= argmax
k
(
R
∑
r=1
δ(Γ(J
ur
) = k)
)
.
where Γ is the relabelling operator performed by the
Hungarian algorithm.
4.3 Bipartite Graph Partitionning
Consensus (BGP)
In this consensus the collection of estimated solutions
is represented by a bipartite graph where the vertices
are divided into two sets: the point vertices and the
label vertices. The point vertices represent the points
of the data matrix {(e
i
, f
j
)} while the set of label ver-
tices represents all the estimated biclusters of the col-
lection {
ˆ
B
k,(r)
}, for each estimated solution there is
also a vertice that represents the points belonging to
no bicluster. An edge links a point vertice to a label
vertice if the point belongs to the corresponding esti-
mated bicluster. The degree of each point is therefore
R and the degree of each estimated bicluster repre-
sents the number of points that it contains. Finding
a consensus consists in finding a partition of this bi-
partite graph. The optimal partition is the one that
maximizes the numbers of edges inside each cluster
of nodes and minimizes the number of edges between
nodes of different clusters. This graph partitioning
problem is a NP-hard problem, so we rely on a heuris-
tic to an approximation of the optimal solution. We
used a method based on a spin-glass model and sim-
ulated annealing (Reichardt and Bornholdt, 2006) in
order to identify the clusters of nodes. Each clus-
ter of the partition represents an aggregated bicluster
formed by all the points contained in this cluster.
4.4 Multivariate Mixture Model
Consensus (MIX)
In (Topchy et al., 2004a), the authors have used the
mixture approach to propose a consensus function. In
the sequel we propose to extend it to our situation.
In model-based clustering it is assumed that the data
are generated by a mixture of underlying probability
distributions, where each component k of the mixture
represents a cluster. Specifically, the NM × R data
matrix is assumed to be an J
¯
1
,...,J
¯
u
,...,J
¯
NM
i.i.d
UnsupervisedConsensusFunctionsAppliedtoEnsembleBiclustering
33
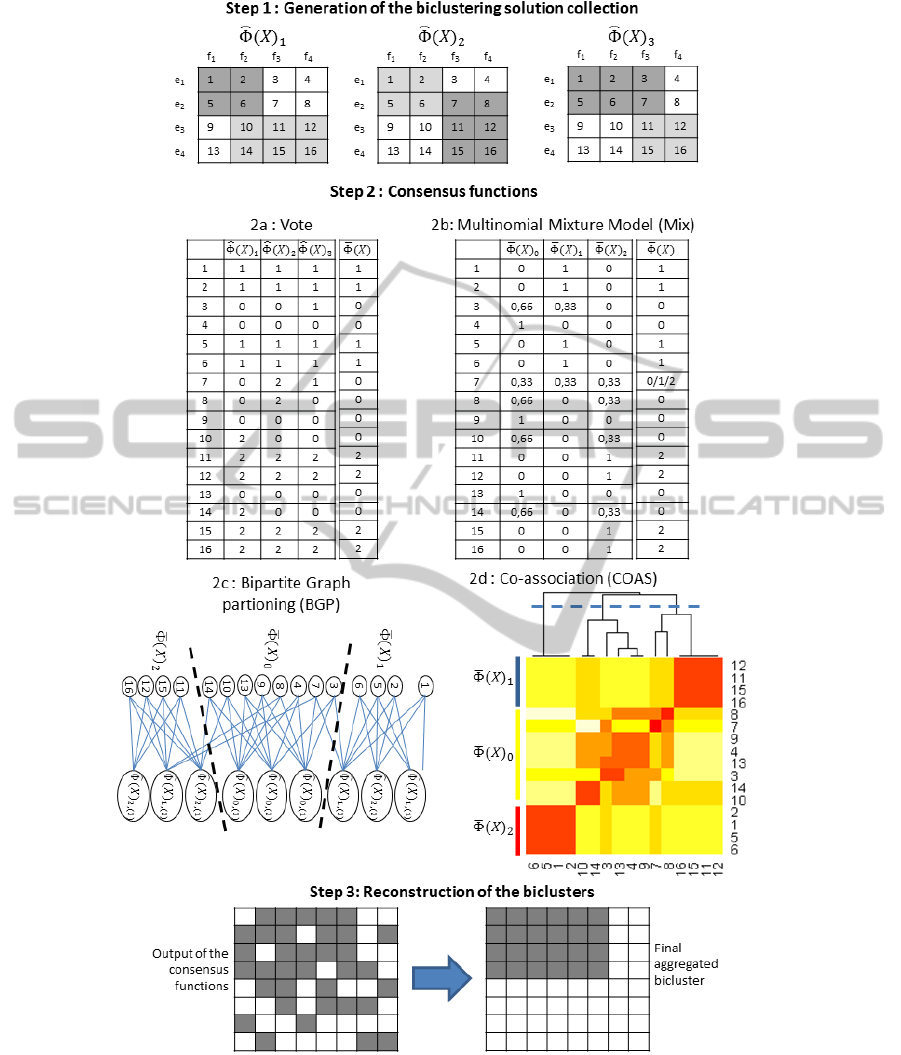
Figure 1: Procedure of ensemble biclustering with the four consensus functions. 1) 3 different biclustering solutions with 2
biclusters for the same data matrix forming the collection. 2a) The collumns represents the labels of each data points obtained
by the three biclustering solution. The last column represents the results of the VOTE consensus. 2b) The first three columns
give the probability for each data point to be associated to the three labels of the mixture model. The last column represents
the results of the MIX consensus. 2c) The bipartite graph representing all biclusters of the collection. The cuts of the graph
give the results of the BGP consensus. 2d) The coassociation matrice of the data points. The 3 clusters obtained from this
matrix represent the results of the COAS consensus. 3) An example of the reconstruction step of our methods.
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
34

sample where J
¯
u
from a probability distribution with
density
φ(J
¯
u
|Θ) =
K
∑
k=0
π
k
P
k
(J
¯
u
|θ
k
),
where P
k
(J
¯
u
|θ
k
) is the density of label J
¯
u
from the kth
component and the θ
k
s are the corresponding class
parameters. These densities belong to the same para-
metric family. The parameter π
k
is the probability that
an object belongs to the kth component, and K, which
is assumed to be known, is the number of components
in the mixture. The number of components corre-
sponds to the number of biclusters minus one since
one of the components represents the points belong-
ing to no bicluster. The parameter of this model is
the vector Θ = (p
¯
i
0
,...,p
¯
i
K
,θ
0
,...,θ
K
). The mixture
density of the observed data can be expressed as
φ( |Θ) =
NM
∏
u=1
K
∑
k=0
π
k
P
k
(J
¯
u
|θ
k
).
The J
¯
u
labels are nominal categorical variables, we
consider the latent class model and assume that all R
categorical variables are independent, conditionnally
on their memebership of a component;
P
k
(J
¯
u
|θ
k
) =
R
∏
r=1
P
k,(r)
(J
ur
|θ
k,(r)
).
Note that P
k,(r)
(J
¯
u
|θ
k,(r)
) represents the probability to
have J
¯
u
labels in the kth component for the estimated
solution
ˆ
Φ(X)
(r)
. If α
r( j)
k
is the probability that the
rth label takes the value j when an J
¯
u
belongs to the
component k, then the probability of the mixture can
be written P
k
(J
¯
u
|θ
k
) =
∏
R
r=1
∏
K
j=1
[α
r( j)
k
]
δ(J
ur
= j)
. The
parameter of the mixture Θ is fitted in maximizing
the likelihood function:
Θ
∗
= argmax
Θ
(
log
(
NM
∏
u=1
P(J
¯
u
|θ)
))
.
The optimal solution of this maximization problem
cannot generally be computed, we therefore rely on
an estimation given by the EM algorithm (Dempster
et al., 1977). In E-step, we compute the posterior
probabilities of each label s
uk
∝ P
k
(J
¯
u
|θ
k
) and in the
M-step we estimate the parameters of the mixture as
follows
π
k
=
∑
u
s
uk
NM
and α
r( j)
k
=
∑
u
s
uk
δ(J
ur
= j)
∑
u
s
uk
.
To limit the problems of local minimum during the
EM algorithm, we performd the optimization process
ten times with different initializations and kept the so-
lution maximizing the log-likelihood. At the conver-
gence, we consider that the largest π
k
corresponds to
S1 S2 S3 S4
Figure 2: The four data structures considered in the experi-
ments.
labels representing the points belonging to no biclus-
ters. The estimators of posterior probabilities give
rise to a fuzzy or hard clustering using the maxi-
mum a posteriori principle (MAP). Then the consen-
sus function consists in taking for each J
¯
u
the clus-
ter such that k maximizing its conditional probability
k = argmax
ℓ=1,...,K
s
uℓ
, and we obtained the ensemble
solution noted Φ( ).
4.5 Reconstruction of the Biclusters
The four consensus functions presented above, return
a partition in K + 1 clusters of the points of the data
matrix. K of these clusters represent the K aggregated
biclusters, the last one groups all the points that be-
long to no biclusters in the aggregated solution. The
k aggregated biclusters are not actual biclusters yet.
They are just sets of points that do not necessarily
form submatrices of the data matrix. A reconstruction
step has to be applied to each aggregated bicluster in
order to transform it into a submatrix. This proce-
dure consists in finding the submatrix containing the
maximum of points that are in the aggregated biclus-
ter and the minimum of points that are not in the ag-
gregated bicluster. The k-th aggregated bicluster is
reconstructed by minimizing the following function:
L(B
k
) =
N
∑
i=1
M
∑
j=1
δ(e
i
∈ E
B
k
∧ f
i
∈ F
B
k
)δ(I
i j
̸= k)
+ δ(e
i
/∈ E
B
k
∨ f
i
/∈ F
B
k
)δ(I
i j
= k).
This optimization problem is solved by a heuristic
procedure. We started with all the examples and fea-
tures involved in the aggregated bicluster. Then iter-
atively, we dropped the example or feature that max-
imizes the decrease of L(B
k
). This step was iterated
until L(B
k
) did not decrease. Once the reconstruction
procedure was finished, we obtained the final aggre-
gated biclusters.
UnsupervisedConsensusFunctionsAppliedtoEnsembleBiclustering
35

5 RESULTS AND DISCUSSION
5.1 Performance of Consensus
Functions
In our simulations, we considered four different data
structures with M = N = 100 in which a true bi-
clustering solution is included. The number of bi-
clusters varies from 2 to 6 and their sizes from 10
examples by 10 features to 30 examples by 30 fea-
tures. We have defined four different structures of
biclusters depicted in Figure 2. For each data, from
each true bicluster an estimated bicluster was gener-
ated, then a collection of estimated biclustering so-
lutions was obtained. The quality of the collection
is controlled by the parameters α
pre
and α
rec
that
are the average precision and recall between esti-
mated biclusters and their corresponding true biclus-
ters. To generate an estimated bicluster we started
with the true bicluster, then we randomly removed
features/examples and have added features/examples
that were not in the true bicluster in order to ob-
tain the target precision α
pre
and recall α
rec
. Once
the collection was generated, the four consensus
functions were applied to obtain the aggregated bi-
clustering solutions. Finally to evaluate the perfor-
mance of each aggregated solution we computed the
average F-measure (noted ∆) between the obtained
solution Φ( ) and the true biclustering solution
Φ( )
∗
; ∆(Φ( )
∗
,Φ( )) =
1
K
∑
K
k=1
M
Fmes
(B
∗
k
,B
k
)
where M
Fmes
(B
∗
k
,B
k
) =
|B
∗
k
∩B
k
|
|B
∗
k
|+|B
k
|
is the F-measure.
Figure 3 shows the performance of the different
consensus in function on the size of the biclustering
solution collection R with α
pre
= α
rec
= 0.5. Each of
the six panels gives the results on the six data struc-
tures. The dot, triangle, cross and diamond curves
represent respectively the F-measure in function of R
for VOTE, COAS, BGP and MIX consensus. The full
gray curve represents the mean of the performance of
the biclustering collection. In the six panels, the per-
formance of the collection is constantly around 0.5.
That is be expected, since the performance of the col-
lection does not depend on its size and by construction
the theoretical performance of each estimated solu-
tion is 0.5. On the six dataset structures, from R ≥ 40,
all the consensus functions give much better perfor-
mances than the estimated solutions of the collec-
tion. The performances of MIX in all the situations
are strongly increasing with the size of the collection.
Mix does not require a high value of R to record good
results, for R ≥ 20 it converges to their maximum and
reaches 1 in all panels. The curves of BGP have the
same shape, they begin with a strong increase then
they converge to their maximums, but the increase
phase is much longer than in MIX. It also worth not-
ing that BGP begins with very low performances for
small values of R, it is often lower than the perfor-
mances of the collection. BGP reaches its best per-
formances with R ≥ 60, in four panels it obtains the
second best results and the third on the two last pan-
els. The performance of VOTE increases slowly and
more or less linearly with the collection size. Even
with very low values of R, the performance of the
consensus is significantly better than the collection.
VOTE gives the second best performances for S1 and
S5 and the third best for the four other data structures.
The performance of COAS is more or less constant
whatever R; it obtains the worst results in all panels.
Figure 4 shows the performances of the different con-
sensus in function of the performances of the esti-
mated solution collection controlled by the parame-
ter α = α
pre
= α
rec
. The performances of all con-
sensus are naturally decreasing with α. By defini-
tion the performances of the collection follow the line
y = 1 − x . For α ≤ 0.4 and in all the cases the con-
sensus functions give the almost perfect biclustering
solution with ∆ ≈ 1, expected for COAS in S4. MIX
is still clearly the best consensus, it produces almost
the perfect biclustering and its performances are never
less than 0.9. BGP is the second best consensus, it is
always significantly better than the collection what-
ever the value of α. VOTE and COAS have simi-
lar behavior. They begin with the perfect bicluster-
ing solution then, when α ≥ 0.5, their performances
decrease and are at best, for VOTE, around the collec-
tion performance.
The F-measure can be decomposed into a combi-
nation of precision and recall. When we examine the
results in detail we see that for VOTE and COAS the
precision is much greater than the recall. That means
these consensus produce smaller biclusters than the
true ones, the features and examples associated to bi-
clusters are generally good but these biclusters are
incomplete i.e. examples and features are missing.
Conversely BGP produces biclusters with high re-
call and low precision. The aggregated biclusters are
generally complete but they also contain some extra
wrong features and examples. MIX gives balanced
biclusters with equal precision and recall. The exper-
iment on S4 makes it possible to observe the influence
of the size of the biclusters on the results. We can see
that COAS obtains very bad performance on the small
biclusters, since the recall on the two smallest biclus-
ters is 0. MIX, VOTE, COAS are independent from
the size of the biclusters, their performances are sim-
ilar with the four biclusters.
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
36
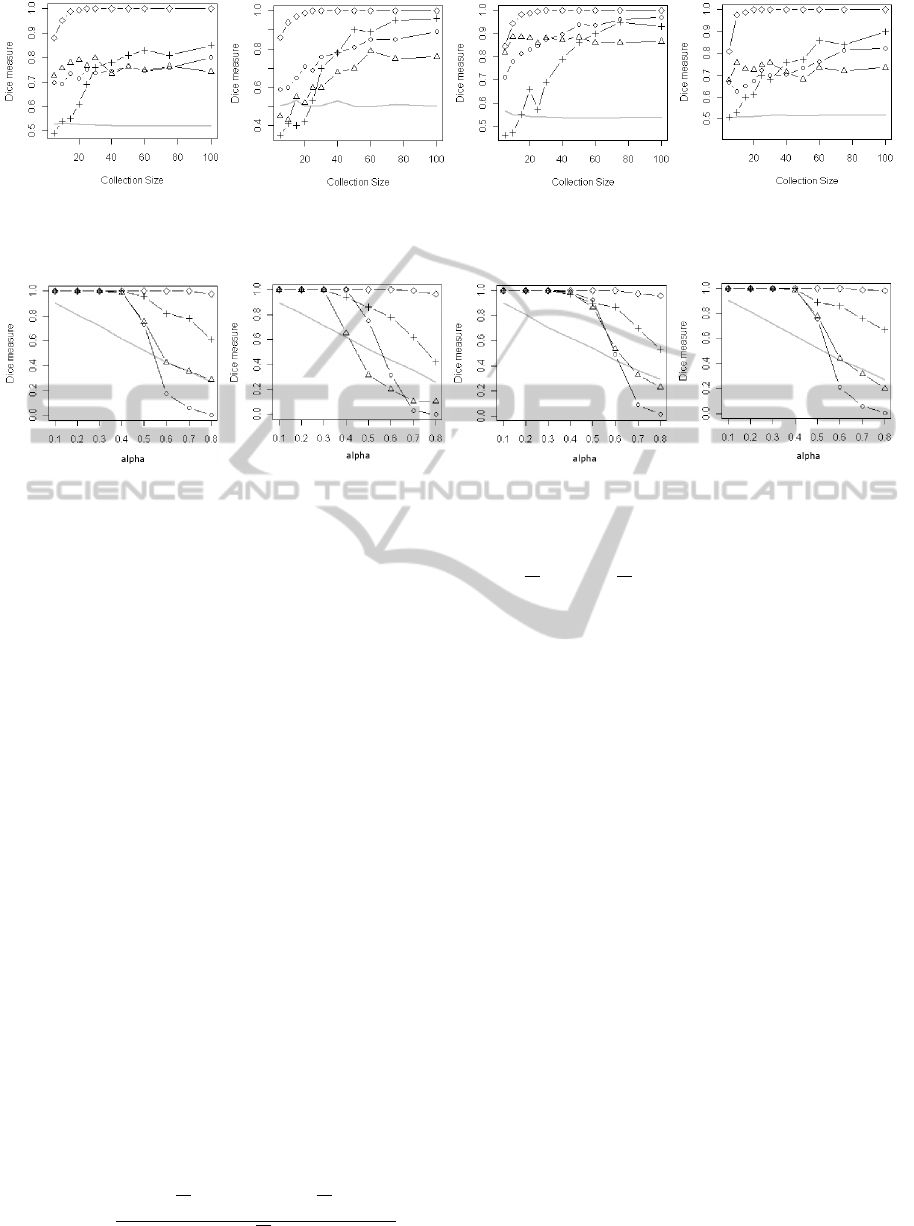
S1 S2 S3 S4
◦ : VOTE, △ : COAS, + : BGP, ♢ : MIX.
Figure 3: Performance of the consensus in function of R (size of the biclustering solution collection) with α
pre
= α
rec
= 0.5.
S1 S2 S3 S4
◦ : VOTE, △ : COAS, + : BGP, ♢ : MIX.
Figure 4: Performance of the consensus in function on the mean precision α
pre
and recall α
rec
of the biclustering solution
collection with α = α
pre
= α
rec
.
5.2 Results on Real Data
To evaluate our approach in terms of performance
on real datasets, we used four datasets: Nutt dataset
(gene expression), Pomeroy dataset (gene expres-
sion), Sonar dataset (Sonar signal) and Wdbc dataset
(biological dataset). Unlike numerical experiments
and since we do not known the true biclustering so-
lutions, the measures of performance can be based on
external indices, like Dice score. Obviously, the qual-
ity of a biclustering solution can be measured by the
AMSR i.e. the average of MSR computed from each
bicluster belonging to the biclustering solution; the
lower the AMSR, the better the solution. A problem
with this approach is that the MSR is biased by the
size of the biclusters. Indeed, the smallest biclusters
favour AMSR. To remove this size bias we set the size
of the biclusters in the parameters of the algorithms.
All the methods will therefore return biclusters of the
same size. The better solutions will be those mini-
mizing AMSR. To compare the different consensus
functions, we computed their gain which is the per-
centage of AMSR decreasing from the single biclus-
tering solution i.e. the solution obtained by the classic
CC algorithm without the ensemble approach. This is
computed by:
Gain = 100
AMSR(Φ
single
) − AMSR(Φ
ensemble
)
AMSR(Φ
single
)
,
where Φ
single
and Φ
ensemble
are the biclustering solu-
tion returned respectively by the single and ensemble
approaches.
Table 1 gives the gain of each consensus function
for all the datasets in function on the size of the bi-
clusters. We can observe that in all the situations, all
the consencus functions give an interesting gain, ex-
pected for COAS for Wdbc dataset. We know that in
the merging process, once a cluster is formed it does
not undo what was previously done; no modifications
or permutations of objects are therefore possible. This
disadvantage can be a handicap for COAS in some sit-
uations such as in Wdbc dataset. VOTE and MIX out-
perform BGP in most cases. In addition their behavior
does not to depend on the size of biclusters. In Nutt
and Sonar datasets, their performance has increased
or decreased respectively. VOTE appears more effi-
cient than MIX for the Nutt dataset which is the larger.
However, the size of the biclusters seems unaffacted
MIX in other experiments. The difference of perfor-
mance between VOTE/MIX and BGP/COAS is large.
We observe that the size of the bicluster may impact
the performance of the methods but there is no clear
rule, it is only dependent on the data. Further inves-
tigation will be necessary. In summary VOTE and
MIX produce the best performances, the third is BGP
and the last is COAS. Knowing that VOTE and MIX
require less computing time than BGP, both appear
therefore more efficient.
UnsupervisedConsensusFunctionsAppliedtoEnsembleBiclustering
37

Table 1: Gain of each consensus function on the four real
datasets in function of the size of the biclusters.
Nutt dataset
50 100 200 300 400 600 800
VOTE 94 64 18 20 34 27 27
MIX 13 3 43 39 36 18 3
COAS 28 37 14 14 32 5 6
BGP 73 68 74 1 30 22 16
Pomeroy dataset
50 100 200 300 400 600 800
VOTE 79 85 79 69 32 63 60
MIX 84 83 69 52 37 75 74
COAS 69 78 21 36 30 43 39
BGP 68 80 21 22 30 46 51
Sonar dataset
50 100 200 300 400 600 800
VOTE 20 30 41 75 93 86 88
MIX 29 47 55 88 92 77 82
COAS 28 17 33 45 72 36 76
BGP 34 51 50 46 20 21 32
Wdbc dataset
50 100 200 300 400 600 800
VOTE 15 20 28 20 4 11 3
MIX 26 19 42 32 23 21 12
COAS -4 -18 -15 -7 -17 -8 -25
BGP 6 13 37 31 2 10 -4
6 CONCLUSIONS
Unlike to the standard clustering contexts, bicluster-
ing considers both dimensions of the matrix in order
to produce homogeneous submatrices. In this work,
we have presented the approach of ensemble biclus-
tering which consists in generating a collection of bi-
clustering solutions then to aggregate them. First, we
have showed how to use the CC algorithm to generate
the collection. Secondly, concerning the aggregation
of the collection of biclustering solutions, we have ex-
tended the use of four consensus functions commonly
used in the clustering context. Thirdly we have eval-
uated the performance of each of them.
On simulated and real datasets, the ensemble ap-
proach appears fruitful. The results show that it im-
proves significantly the performance of biclustering
whatever the consensus function among VOTE, MIX
and BGP. Specifically, VOTE and MIX give clearly
the best results in all experiments and require less
computing than BGP. We thus recommend to use one
of these two methods for ensemble biclustering prob-
lems. For the moment our methods do not allow the
overlapping between examples. However the limita-
tion comes to the implementation of the collection of
the biclusters, the consensus functions are compatible
with the overlapping. The overlapping problem will
be handled in future works.
REFERENCES
Breiman, L. (1996). Bagging predictors. Machine Learn-
ing, 24:123–140.
Breiman, L. (2001). Random forests. Machine Learning,
45:5–32.
Busygin, S., Prokopyev, O., and Pardalos, P. (2008). Bi-
clustering in data mining. Computers and Operations
Research, 35(9):2964–2987.
Cheng, Y. and Church, G. M. (2000). Biclustering of ex-
pression data. Proc Int Conf Intell Syst Mol Biol,
8:93–103.
De Smet, R. and Marchal, K. (2011). An ensemble
biclustering approach for querying gene expression
compendia with experimental lists. Bioinformatics,
27(14):1948–1956.
Dempster, A., Laird, N., and Rubin, D. (1977). Maxi-
mum likelihood from incomplete data via the em al-
gorithm. Journal of the royal statistical society, series
B, 39(1):1–38.
Dhillon, I. S. (2001). Co-clustering documents and words
using bipartite spectral graph partitioning. In Pro-
ceedings of the seventh ACM SIGKDD international
conference on Knowledge discovery and data mining,
KDD ’01, pages 269–274.
Diaz-Uriarte, R. and Alvarez de Andres, S. (2006). Gene
selection and classification of microarray data using
random forest. BMC Bioinformatics, 7(3).
Dietterich, T. G. (2000). Ensemble methods in machine
learning. Lecture Notes in Computer Science, 1857:1–
15.
Dudoit, S. and Fridlyand, J. (2003). Bagging to improve the
accuracy of a clustering procedure. Bioinformatics,
19(9):1090–1099.
Erten, C. and S
¨
ozdinler, M. (2010). Improving perfor-
mances of suboptimal greedy iterative biclustering
heuristics via localization. Bioinformatics, 26:2594–
2600.
Fern, X. Z. and Brodley, C. E. (2004). Solving cluster en-
semble problems by bipartite graph partitioning. In
Proceedings of the twenty-first international confer-
ence on Machine learning, ICML ’04, pages 36–.
Frossyniotis, D., Likas, A., and Stafylopatis, A. (2004). A
clustering method based on boosting. Pattern Recog-
nition Letters, 25:641–654.
Govaert, G. (1995). Simultaneous clustering of rows and
columns. Control and Cybernetics, 24(4):437–458.
Govaert, G. and Nadif, M. (2003). Clustering with block
mixture models. Pattern Recognition, 36:463–473.
Hanczar, B. and Nadif, M. (2010). Bagging for biclustering:
Application to microarray data. In European Confer-
ence on Machine Learning, volume 1, pages 490–505.
Hanczar, B. and Nadif, M. (2012). Ensemble methods for
biclustering tasks. Pattern Recognition, 45(11):3938–
3949.
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
38

Hartigan, J. A. (1972). Direct clustering of a data ma-
trix. Journal of the American Statistical Association,
67(337):123–129.
Lazzeroni, L. and Owen, A. (2000). Plaid models for gene
expression data. Technical report, Stanford Univer-
sity.
Maclin, R. (1997). An empirical evaluation of bagging
and boosting. In In Proceedings of the Fourteenth
National Conference on Artificial Intelligence, pages
546–551. AAAI Press.
Madeira, S. C. and Oliveira, A. L. (2004). Bicluster-
ing algorithms for biological data analysis: a survey.
IEEE/ACM Transactions on Computational Biology
and Bioinformatics, 1(1):24–45.
Papadimitriou, C. H. and Steiglitz, K. (1982). Com-
binatorial optimization: algorithms and complexity.
Prentice-Hall, Inc., Upper Saddle River, NJ, USA.
Reichardt, J. and Bornholdt, S. (2006). Statistical mechan-
ics of community detection. Phys. Rev. E, 74:016110.
Schapire, R. (2003). The boosting approach to machine
learning: An overview. in Nonlinear Estimation and
Classification, Springer.
Strehl, A. and Ghosh, J. (2002). Cluster ensembles - a
knowledge reuse framework for combining multiple
partitions. Journal of Machine Learning Research,
3:583–617.
Topchy, A., Jain, A. K., and Punch, W. (2004a). A mixture
model of clustering ensembles. In Proc. SIAM Intl.
Conf. on Data Mining.
Topchy, A. P., Law, M. H. C., Jain, A. K., and Fred, A. L.
(2004b). Analysis of consensus partition in cluster en-
semble. In Fourth IEEE International Conference on
Data Mining., pages 225–232.
Turner, H., Bailey, T., and Krzanowski, W. (2005). Im-
proved biclustering of microarray data demonstrated
through systematic performance tests. Computational
Statistics & Data Analysis, 48(2):235–254.
van der Laan, M., Pollard, K., and Bryan, J. (2003). A new
partitioning around medoids algorithm. Journal of
Statistical Computation and Simulation, 73(8):575–
584.
UnsupervisedConsensusFunctionsAppliedtoEnsembleBiclustering
39
