
Human Genome in a Smart Card
Mete Akg
¨
un, Bekir Erg
¨
uner, A. Osman Bayrak and M. S¸amil Sa
˘
gıro
˘
glu
T
¨
UB
˙
ITAK B
˙
ILGEM UKEAE (National Research Institute of Electronics and Cryptology), 41470, Gebze, Kocaeli, Turkey
Keywords:
Human Genome, Smart card, Disease Risk Test.
Abstract:
Gene sequencing costs have fallen considerably with advancements in technology in the last 10 years. This
cost will be reduced even further in the following years. That means personal genomics will be very possible
in the near future. How and where genetic information is stored is the biggest problem for individuals. Fur-
thermore, privacy of genomic data has a great importance because it can be used to identify an individual and it
contains privacy-sensitive data. Therefore, privacy-preserving methods for the use of genomic data should be
developed. In this paper, we present a method for storing some parts of human genome needed for the disease
risk computation in a smart card. Our method uses variations that separate any genome from the reference
genome. It selects the variations corresponding to the exonic regions and filters them according to their variant
quality and genotype quality. We show that our method can reduce a single whole human genome to the size
small enough to be stored in a smart card without abusing the genomic privacy of individuals. Furthermore,
we also propose a simple system in which genomic smart cards are used to perform privacy preserving disease
risk test.
1 INTRODUCTION
Next Generation Sequencing technologies are paving
the way to individual genomics and personalized
medicine. In the near future, decline in the cost of
genome sequencing would break the bond between
doctor and patient. That means every person will want
to have their own genome without contacting doctors.
Storage and retrieval of personal genomes would ap-
pear to be a major problem. Gene sequences provide
information about the current health status of people
they belong to. In addition, they carry information
about the health problems that were experienced in
the past and may occur in the future. Therefore, the
privacy of gene sequences is of great importance.
High-throughput sequencing methodologies are
capable of producing large amount of sequencing
data. This data may occupy tens or even hundreds
of gigabytes of disk space. Although individual ge-
nomics is not common, the storage of these large
amount data is already a big problem for researchers
and clinicians. Genomic science shows the disposi-
tion of a person to any disease stems from variations
in its genome. Clinicians can use genomic variations
for diagnosis and preventive medicine. Genomic data
provides opportunities for substantial improvements
in diagnosis and preventive medicine. In particular, it
has been shown that an individuals predisposition to
a disease depends on genomic variations. There are
some companies such as 23andMe (23andme, 2013)
and Counsyl (Counsyl, 2013) that offer the calcula-
tion of genetic risk for some diseases.
Containing private and sensitive personal data
such as genetic predisposition to certain diseases, in-
dividual genomic data attracts broad interest of a large
variety of health-care stakeholders like pharmaceuti-
cal and private insurance companies, physicians and
on line direct-to-consumer service providers. To pro-
tect the privacy of his genomic data, an individual
might not want to disclose this sensitive data to other
parties. For example, while getting a risk assessment
service for a specific disease from a medical unit, a
patient should be able not to reveal any further infor-
mation apart from that specific disease to either the
medical unit or the physician. The only information
that the medical unit or the physician might get would
be the risk level. Thus, for accuracy of genetic pre-
disposition tests, protecting genomic data privacy of
individuals is crucial as well as using the correct and
complete data.
In this paper, we present a method for storing im-
portant parts of human genome needed for the dis-
ease risk computation in a smart card. Our method
uses variations that separate any genome from the ref-
erence genome. It selects the variations correspond-
ing to the exonic regions and filters them according to
310
Akgün M., Ergüner B., Bayrak A. and Sa
˘
gıro
˘
glu M..
Human Genome in a Smart Card.
DOI: 10.5220/0004799903100316
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2014), pages 310-316
ISBN: 978-989-758-010-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

their variant quality and genotype quality. It indexes
the previously known variations by using by using
Single Nucleotide Polymorphism (dbSNP) database
(Sherry et al., 2001). Furthermore, our method deals
with the variations that are specific to the person. We
show that our method can reduce a single whole hu-
man genome to the size small enough to be stored in
a smart card. We also propose a system in which ge-
nomic smart cards are used to perform privacy pre-
serving disease risk test.
2 RELATED WORK
In (Reber and Perttunen, 1997), a method storing at
least a portion of human genome on on the machine-
readable storage medium was proposed. This method
proposes the storage of raw genome in a lossless man-
ner. However, the storage limits and the data process-
ing method are not specified.
In the literature, there are many solutions for pro-
tecting the privacy of genomic data used in genetic
tests. Troncoso-Pastoriza et al. (Troncoso-Pastoriza
et al., 2007) proposed a protocol for secure pat-
tern matching by evaluating automata in an oblivious
manner. The protocol is developed for secure DNA
matching and provides security in semi-honest set-
ting. The communication complexity is linear in the
size of input alphabet and the number of states of the
finite state machine.
Adida and Kohane (Adida and Kohane, 2006) pro-
posed a system GenePING for secure storage of large,
genome-sized datasets. GenePING is developed by
extending the PING (Riva et al., 2001; Simons et al.,
2005) personal health record system. The authors
claim that an attacker accessing to the raw GenePING
storage can not find any relation between patient and
genomic data points.
Blanton and Aliasgari (Blanton and Aliasgari,
2010) proposed a solution for secure DNA searching
also using secure automata evaluation. Proposed pro-
tocol introduce improvements on (Troncoso-Pastoriza
et al., 2007) reducing communicating parties work.
This work proposes a secure outsourcing of computa-
tion protocol which uses external service provider and
modified multi-party protocol.
Jha et al. (Jha et al., 2008) presents privacy
protecting implementations on genomic computations
such as sequence comparisons and distance calcula-
tions using secure two-party communication proto-
col. For edit distance they developed three protocols.
First protocol uses Yao’s garbled circuits while sec-
ond combines garbled circuits with secure computa-
tion with shares. To overcome performance and scal-
ability issues they hybridized the first two protocols
into the third.
Bruekers et al. (Bruekers et al., 2008) proposed
a solution, in semi-honest attacker model, for limited
DNA-based operations like identity, ancestor and pa-
ternity tests, based on Short Tandem Repeat. Pro-
vided solution is based on homomorphic encryption
and its complexity highly depends on the number of
errors to be tolerated.
Taking into account the fully-sequenced human
genome, Baldi et al. (Baldi et al., 2011) proposed
protocols, for paternity tests, personalized medicine
and genetic compatibility tests, based on private set
operations technique. The main aim of this work is to
provide privacy protecting mechanisms to individu-
als, who have their genomic data, for getting serviced
for genetic tests from authorized parties.
Canim et al. (Canim et al., 2012) proposed to
use cryptographic hardware for secure storage, share
and query of genomic data. As a tamper-proof hard-
ware, secure coprocessors are employed for process-
ing genomic data owned by health organizations e.g.
hospitals. Data reside, in encrypted form, in data
storage servers which are assumed to be untrusted.
Proposed solution only addresses the genomic data
owned by health organizations and use potentially ex-
pensive tamper-proof hardware. As the authors agree,
the proposed protocol cannot provide privacy in the
case where information is extracted from the query
results.
Ayday et al. (Ayday et al., 2012) proposed a pri-
vacy protecting method, for medical tests and person-
alized medicine using genomic data, based on homo-
morphic encryption. The authors evaluate that per-
sonal genomic data is quite sensitive to be left to in-
dividuals own and propose to store personal genomic
data, in encrypted format, in a storage and process-
ing unit which can be in the control of governments,
non-profit organizations or private companies, such as
cloud storage service providers.
Focusing on disease risk tests, Ayday et al. (Ayday
et al., 2013) proposed a system to provide privacy-
protecting methods, based on homomorphic encryp-
tion, for genomic, clinical and environmental data
storage and process. Like (Ayday et al., 2012), this
work also proposes to use storage and processing unit
to store sensitive data in encrypted form and disease
risk tests are performed by authorized institutions us-
ing homomorphic encryption technique and secure in-
teger comparison.
HumanGenomeinaSmartCard
311

3 BACKGROUND
3.1 Smart Health Cards
Worldwide, some health care organizations imple-
mented smart card applications to store and track
heath records of patients. These applications have
some advantages over traditional paper-based or
computer-based systems:
• More security and privacy for patient data
• Less fraud in healthcare
• Secure transfer of medical records
• Secure access to medical records in case of emer-
gency
• Secure platform for implementing other health-
care applications
• Secure computation module
• Interoperability
• Low implementation cost
• Modular solution
In healthcare applications, the major issue is not
the conversion of traditional health records to elec-
tronic health records (EHRs). Security and the tech-
nology used raises additional problems for healthcare
applications. The format of electronic health records
should be standardized, readable and usable. Elec-
tronic health records should be kept secure, private,
shareable with healthcare stakeholders and easily ac-
cessible in case of emergency. Smart card technology
can meet these requirements of EHR-based health-
care applications. Furthermore, smart card technol-
ogy provides secure computation facility. Smart cards
can be used for secure calculation of some basic op-
erations. Thus, some basic analyses can be done on
smart cards by using electronic health records.
3.2 Variant Call Format (VCF)
Variant Call Format (VCF) (Danecek et al., 2011) is a
text file format storing gene sequence variations such
as SNPs, insertions, deletions and structural variants.
The major advantage of VCF is that only variations
are stored with annotations and positions on the ref-
erence genome. BCF (Binary VCF) format is binary
version of VCF format. The type of information kept
in each format is essentially the same. Supporting
both BCF and VCF file is just a matter of software
update. It does not concern the main goal of our study.
Variant Call Format has three parts. Meta-
information part contains lines beginning with ”##”
and giving the description of FORMAT, INFO and
FILTER entries. There are some optional lines such
as GT and GP in meta-information part. VCF file has
one header line beginning with ”#CHROM”. In VCF
file, each variation is represented in one tab-delimited
data line. A data line is called a VCF record. The first
nine fields of the record are used to describe variants.
• CHROM: an identifier from the reference
genome.
• POS: position in the reference genome.
• ID: A unique identifier for variant (rs number). If
a variation is not pre-defined, the missing value is
used
• REF: reference bases (A,C,G,T,N).
• ALT: comma separated list of alternate non-
reference alleles.
• QUAL: phred-scaled quality score for the asser-
tion made in ALT.
• FILTER: PASS if this position has passed all fil-
ters. Otherwise, a semicolon-separated list of
codes for filters that fail.
• INFO: additional information. As with the INFO
field, there are several common, reserved key-
words that are standards across the community
such as GQ (conditional genotype quality, en-
coded as a phred quality) and DP (read depth at
this position for this sample).
• FORMAT: colon-separated list of data subfields.
4 PERSONAL GENOMIC SMART
CARDS
In this study, we propose usage of smart cards for se-
cure storage of personal genomes. Nowadays, smart
cards are used as an ID card in many countries. Fur-
thermore, there are smart health card applications in
many countries such as France (Smart Card Alliance,
2006a), Germany (Smart Card Alliance, 2006b) and
Taiwan (Smart Card Alliance, 2005). If personal
genomic data is considered as an electronic health
record, our proposal can be applied to existing appli-
cations easily.
The human genome consists of 3 billion base
pairs. Each pair can be represented with two bits.
That means the entire human genome occupies ap-
proximately 750 Megabytes of disk space. At least
99% of the human genome sequence is the same in
all people. Therefore, there is a difference of at most
1% between any human genome and the reference hu-
man genome. Any human genome can be represented
with variations from the reference genome. Typically,
HEALTHINF2014-InternationalConferenceonHealthInformatics
312

Figure 1: Variant Call Format (Image taken from (Wikipedia, 2013)).
these variations are stored in VCF format. The size
of VCF file is usually more than 1 GB that is too big
to store in a smart card. Therefore, we need an extra
method to represent human genome in a smart card.
4.1 Creating Index File
The Single Nucleotide Polymorphism Database
(Sherry et al., 2001) (dbSNP) is a free public archive
for genetic variation within and across different
species developed and hosted by the National Center
for Biotechnology Information (NCBI) in collabora-
tion with the National Human Genome Research In-
stitute (NHGRI). In this study, we work on the dbSNP
build 137 for Homo sapiens (dbsnp 137.b37.vcf). It
contains a range of molecular variation: (1) SNPs,
(2) short deletion and insertion polymorphisms (in-
dels/DIPs), (3) microsatellite markers or short tandem
repeats (STRs), (4) multinucleotide polymorphisms
(MNPs), (5) heterozygous sequences, and (6) named
variants.
Variations which alter the protein coding ex-
onic regions have the most direct effect on pheno-
typic properties. Most of the high penetrance ge-
netic diseases are associated with exonic variants.
Therefore we select the variations which are in ex-
onic regions, decreasing the amount of stored data
by 100 fold while keeping most of the genetic in-
formation. TruSeq Exome Targeted Regions BED
file (Illumina, ) is a file containing information on
the Targeted Exons in the TruSeq Exome product.
We get the intersection of dbsnp 137.b37.vcf and
TruSeq exome targeted regions.b37.bed files with
the help of the intersectBed utility of bedtools (Quin-
lan and Hall, 2010). In this way, we obtain the list
of predefined variations corresponding to exonic re-
gions.
The primary goal of our study is to store maxi-
mum amount of functional information in minimum
amount of storage space. This is crucial for making it
cost effective to apply this method to common clini-
cal use. It is evident that protein coding regions con-
tain the most valuable functional information and the
weight of functional value of non-coding regions is
unclear. Moreover, whole exome sequencing meth-
ods are gaining popularity fast, so exonic variations
are becoming more accessible. We opt to keep only
exonic variations in smart cards. Still, it is possible
to add the variations in regions which have clinical
evidence for their functional value to the reference
database. This way it becomes possible to store the
most valuable information in a smart card.
4.2 Representing a Human Genome in a
Smart Card
In this study, we represent a human genome in
a smart card with variations from the reference
genome. As we mention above, we generate the
list of variations corresponding to the exonic re-
gions by intersecting the corresponding VCF file with
TruSeq exome targeted regions.b37.bed file. We sort
the list of variants according to ID field.
We filter variations according to FILTER field,
conditional genotype quality (GQ) field and read
depth (DP) field. The variation is selected, If FILTER
= PASS (meaning the variation has passed all filters)
and DP ≥ 8 and GQ ≥ 15.
In the previous subsection, we explain how we
create the index file. For each variation in the index
file, we reserve 1-bit field from the storage of smart
card. If the VCF file that will be indexed includes the
corresponding variation, 1-bit field is set to 1 other-
wise it is set to 0. If the number of variations in the in-
dex file is not the multiple of 8, we pad this data with
the required number of bits. We use Huffman Cod-
ing in order to decrease the size of data. We choose
Huffman Coding because a bit in any position can be
obtained without extracting all data.
HumanGenomeinaSmartCard
313
A human genome can include variations that are
not defined in dbSNP. We store these variations in the
format shown in Table 1 and Table 2
Table 1: Variation Storage Format.
Type Genotype Position Information
2 bits 1 bit 32 bits
∗
∗
Please see Table 2
Table 2: Allocation of Information Field.
Type The Number of Bits in
Information Field
SNP (0) 2 bits for reference base
Deletion (1) 16 bits for the size of
deletion
Insertion (2) (16 bits +
(SizeOfInsertion * 2 bits))
for the size of insertion
and DNA sequence
4.3 A Simple System for Disease Risk
Computation
In (Ayday et al., 2013), Ayday et al. proposed a secure
system for privacy preserving computation of the dis-
ease risk. In this system, all genomic data (SNP con-
tent) are stored in Secure Processing Unit (SPU). The
genetic test is performed by using homomorphic en-
cryption and privacy preserving integer comparison.
The effect of each SNP to a disease is expressed with
odds ratio (OR). The OR is the ratio of the proportion
of individuals in the case group having a specific ge-
netic variation to the proportion of individuals in the
control group having the same genetic variation. If
a disease is related with multiple variations, the dis-
ease risk is calculated by taking the weighted aver-
age of the OR of each related variations. Ayday et
al. used logistic regression model in Equation 1 for
disease risk computation.
ln(
Pr
1 − Pr
) = α +
∑
i
β
i
X
i
(1)
They calculate OR of SNP
i
as a OR
i
= exp(β
i
) where
β
i
is the regression coefficient. The calculation over-
all genetic risk is shown in Equation 2.
ln(
Pr
g
1 − Pr
g
) = α +
∑
iεϕx
β
i
p
i
j
(X) (2)
In Equation 2, the effect of the SNP
i
to the overall ge-
netic risk is represented with p
i
j
and α is the intercept
of the model.
In this study, we propose a simple system in which
genomic smart cards can be used for privacy preserv-
ing risk computation. The proposed system is sum-
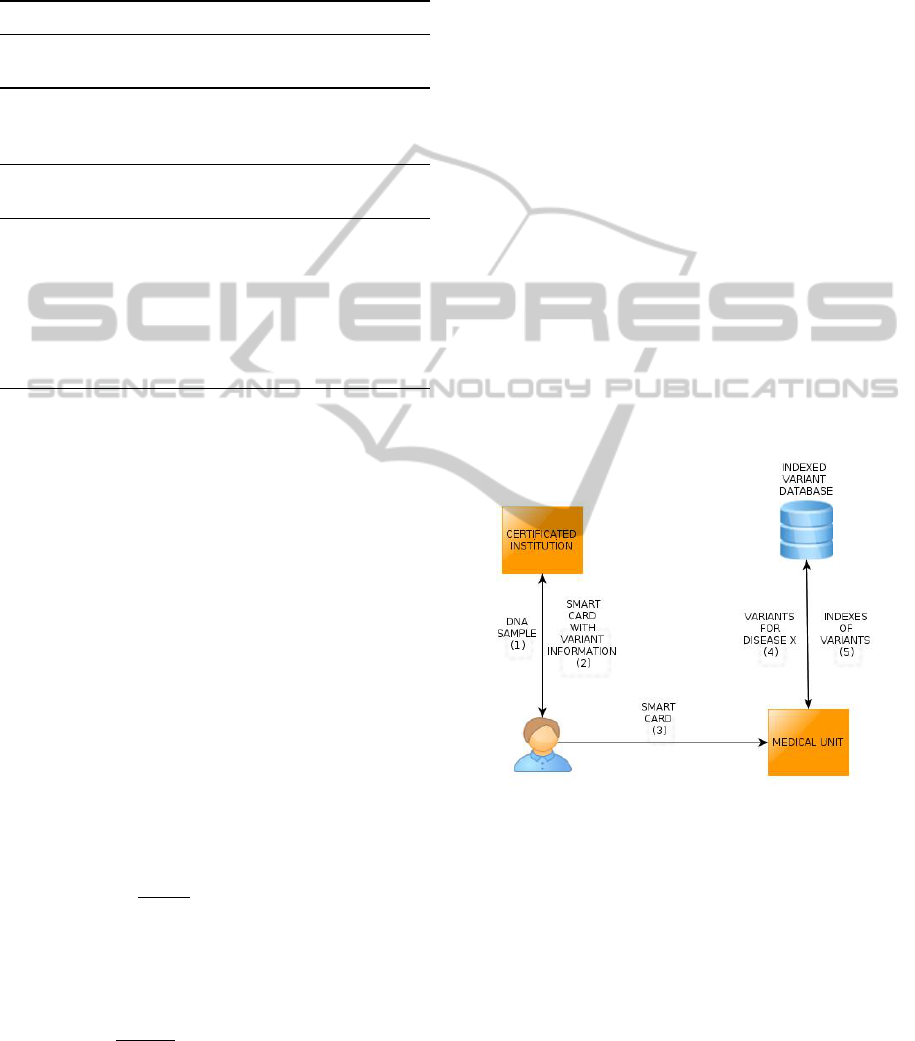
marized in Figure 2. In Step 1, an individual provides
his DNA sample to the certificated institution (CI) for
genome sequencing. CI generates a smart card con-
taining genomic data that is created with the method
explained in Section 4.1 and 4.2. In Step 2, CI sends
the genomic smart card to the individual. The individ-
ual wants to check his disease susceptibility for dis-
ease X. In Step 3, the individual provides his genomic
smart card to the medical unit (MU) for susceptibility
test. MU has access to indexed variant database (IVD)
that is generated as described in Section 4.1. IVD is
stored locally or is accessed over the remote server.
In Step 4, MU queries IVD for variants that poten-
tially cause the disease X. In Step 5, MU gets the in-
dexes of queried variants. In this point, MU sends in-
dexes of corresponding variants and contribution co-
efficients of corresponding variants to the smart card.
The smart card checks whether the individual has the
corresponding variants and computes the disease risk.
Risk calculations can be done with the method used
in (Ayday et al., 2013).
Figure 2: Usage of Genomic Smart Cards.
In this system, we do not need homomorphic en-
cryption and privacy preserving integer comparison.
We know smart cards can be equipped with secure
crypto processors. These microprocessors are tamper-
resistant so they can be used to store and process pri-
vate or sensitive information. The information on
smart card is not accessible through external means
and can be accessed only by the embedded software
which should contain the appropriate security mea-
sures. In our system, an attacker or MU can not obtain
the contents of variants because disease risk compu-
tation is performed in secure crypto processor of the
smart card. Therefore, our method preserves the pri-
HEALTHINF2014-InternationalConferenceonHealthInformatics
314

vacy of genomic data relying on the security strength
of secure crypto processor.
As smart cards are tamper-proof devices, after
writing the genomic data, it will be marked as secret
and will not be readable from out-side the card. That
means, like secret and private keys, genomic data will
not be disclosed, at any time of the card’s lifetime,
by the card once it is written. Only some functions
which will be executed by card’s microprocessor, will
be available for computing disease risks. These func-
tions will take SNP IDs and their risk factors as pa-
rameters and will return a disease risk e.g. as a per-
centage or score. To prevent queries without consent
of the card holder, at every call to these functions,
card will ask its owner to enter his/her PIN. By this
method, even two consecutive queries will require the
card holder to enter his/her PIN twice, therefore a
function call can not be made without holder’s per-
mission.
5 EXPERIMENTAL RESULTS
We implemented our method in C language. In our
experiment, we use six whole genomes VCF files.
These VCF files are obtained from the whole genome
sequencing that are done by Advanced Genomics
and Bioinformatics Research Group (
˙
IGBAM) of In-
formatics and Information Security Research Center
(T
¨
UB
˙
ITAK B
˙
ILGEM).
Table 3: Data Sizes on a Smart Card.
Sample VCF
File
(KByte)
Compressed
Data
(Byte)
Undefined
Varia-
tions
(Byte)
Total
(Byte)
1 1.188.364 73.080 9.487 82.567
2 1.151.748 72.484 8.833 81.317
3 1.146.093 71.978 7.955 79.933
4 1.174.939 72.563 8.747 81.310
5 1.304.739 70.587 3.626 74.213
6 1.237.843 71.144 12.129 83.273
Table 3 shows the results for six different whole
genomes. A typical smart card holds 256 kilobytes
of data and new high capacity cards hold 4 to 256
megabytes without compromising security. Our ex-
periment shows that our method requires maximum
100 kilobytes space. That means it is feasible for low
capacity cards and it can be easily implemented for a
whole human genome.
6 CONCLUSIONS
In this paper, we present a method for storing hu-
man genome in a smart card. Any human genome
can be represented with variations from the reference
genome. Our method selects the variations corre-
sponding to the exonic regions and filters them ac-
cording to their variant quality and genotype qual-
ity. It indexes the previously known variations by us-
ing dbSNP database. Furthermore, our method deals
with the variations that are specific to the person. We
show that our method can reduce a single whole hu-
man genome to the size small enough to be stored in a
smart card. We also present a privacy preserving sys-
tem in which genomic smart cards are used for disease
risk computation. There are two important advantages
of the proposed system: no need for infrastructure and
no need to operate on encrypted data.
As a future work, we will implement our method
on a Java Card smart cards. Thus we can compare the
performance of our disease risk computation method
with those of previously proposed methods.
REFERENCES
23andme (2013). https://www.23andme.com/welcome/.
[Online; accessed 16-July-2013].
Adida, B. and Kohane, I. (2006). Geneping: secure, scal-
able management of personal genomic data. BMC Ge-
nomics, 7(1):1–10.
Ayday, E., Raisaro, J. L., and Hubaux, J.-P. (2012). Privacy-
Enhancing Technologies for Medical Tests Using Ge-
nomic Data. Technical report.
Ayday, E., Raisaro, J. L., Laren, M., Jack, P., Fellay, J., and
Hubaux, J.-P. (2013). Privacy-Preserving Computa-
tion of Disease Risk by Using Genomic, Clinical, and
Environmental Data. In Proceedings of USENIX Se-
curity Workshop on Health Information Technologies
(HealthTech ’13).
Baldi, P., Baronio, R., De Cristofaro, E., Gasti, P., and
Tsudik, G. (2011). Countering gattaca: efficient and
secure testing of fully-sequenced human genomes. In
Proceedings of the 18th ACM conference on Com-
puter and communications security, CCS ’11, pages
691–702, New York, NY, USA. ACM.
Blanton, M. and Aliasgari, M. (2010). Secure outsourcing
of dna searching via finite automata. In Proceedings
of the 24th annual IFIP WG 11.3 working conference
on Data and applications security and privacy, DB-
Sec’10, pages 49–64, Berlin, Heidelberg. Springer-
Verlag.
Bruekers, F., Katzenbeisser, S., Kursawe, K., and Tuyls,
P. (2008). Privacy-preserving matching of dna pro-
files. Cryptology ePrint Archive, Report 2008/203.
http://eprint.iacr.org/.
HumanGenomeinaSmartCard
315

Canim, M., Kantarcioglu, M., and Malin, B. (2012). Secure
management of biomedical data with cryptographic
hardware. Trans. Info. Tech. Biomed., 16(1):166–175.
Counsyl (2013). https://www.counsyl.com. [Online; ac-
cessed 16-July-2013].
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks,
E., DePristo, M. A., Handsaker, R. E., Lunter, G.,
Marth, G. T., Sherry, S. T., McVean, G., Durbin, R.,
and Group, . G. P. A. (2011). The variant call format
and vcftools. Bioinformatics, 27(15):2156–2158.
Illumina. Truseq exome targeted regions bed file.
http://support.illumina.com/downloads/truseq exome
targeted regions bed file.ilmn. [Online; accessed
19-December-2013].
Jha, S., Kruger, L., and Shmatikov, V. (2008). Towards
practical privacy for genomic computation. In Secu-
rity and Privacy, 2008. SP 2008. IEEE Symposium on,
pages 216–230.
Quinlan, A. R. and Hall, I. M. (2010). Bedtools: a flexi-
ble suite of utilities for comparing genomic features.
Bioinformatics, 26(6):841–842.
Reber, William, L. and Perttunen, Cary, D. (1997). Personal
human genome card and methods and systems for pro-
ducing same. Patent Application. WO 1997/031327
A1.
Riva, A., Mandl, K. D., Oh, D. H., Nigrin, D. J., Butte, A. J.,
Szolovits, P., and Kohane, I. S. (2001). The personal
internetworked notary and guardian. I. J. Medical In-
formatics, 62(1):27–40.
Sherry, S. T., Ward, M., Kholodov, M., Baker, J., Phan, L.,
Smigielski, E. M., and Sirotkin, K. (2001). dbsnp:
the ncbi database of genetic variation. Nucleic Acids
Research, 29(1):308–311.
Simons, W. W., Mandl, K. D., and Kohane, I. S. (2005).
Model formulation: The ping personally controlled
electronic medical record system: Technical architec-
ture. JAMIA, 12(1):47–54.
Smart Card Alliance (2005). Taiwan
health care smart card project. http://
www.smartcardalliance.org/resources/pdf/Taiwan
Health Card Profile.pdf”. Accessed: 2013-11-25.
Smart Card Alliance (2006a). French
sesam vitale health card. http://
www.smartcardalliance.org/resources/pdf/Sesam
Vitale.pdf. Accessed: 2013-11-25.
Smart Card Alliance (2006b). German health card. http://
www.smartcardalliance.org/resources/pdf/German
Health Card.pdf. Accessed: 2013-11-25.
Troncoso-Pastoriza, J. R., Katzenbeisser, S., and Celik, M.
(2007). Privacy preserving error resilient dna search-
ing through oblivious automata. In Proceedings of the
14th ACM conference on Computer and communica-
tions security, CCS ’07, pages 519–528, New York,
NY, USA. ACM.
Wikipedia (2013). Variant call format
— Wikipedia, the free encyclopedia.
https://en.wikipedia.org/wiki/Variant Call Format.
[Online; accessed 8-July-2013].
HEALTHINF2014-InternationalConferenceonHealthInformatics
316
