
BioMetaDB: Ontology-based Classification and Extension of
Biodatabases
Ching-Fen Chang, Chang-Hsien Lin and Chuan-Hsiung Chang
Institute of Biomedical Informatics, Center for Systems and Synthetic Biology, National Yang Ming University,
Taipei 11221, Taiwan
Keywords: Information Retrieval, Ontology, Classification, Biodatabase, Semantics, Corpus.
Abstract: The recent rapid increase in high-throughput biological data and computational tools has facilitated the
establishments of numerous biodatabases as the repositories of biodata and bioinformatics analysis tools.
Due to the inefficiency of database categorization, the search of all available information of research
interests costs researchers a lot of time and efforts. We have established BioMetaDB for users to
systematically identify all the available databases of their interests and to extend databases on relevance
biomedical contents. For the purpose of establishing BioMetaDB to provide semantically annotated corpus
to markup the instances of biomedical ontology, our BioMetaDB comprises three main tasks: (1) biological
information retrieval from public databases; (2) creating an integrated ontology repository for biological and
medical studies based on expert-tagged corpus; (3) establish web services to enable users to access all their
desired databases by systemically ontology query. Based on biomedical ontologies, we indexed all the
databases by their relevant biological features, and further evaluated the relevance among the databases. Our
BioMetaDB, a comprehensive compendium of biological databases, is currently integrated from over 1,500
digital sources.
1 INTRODUCTION
The growing amounts of biomedical databases, high-
throughput biological data and computational tools
are driving the need for more effective methods for
database indexing, searching, and understanding.
However, database content detection based on
classification technique is still a challenging
research issue. The search of all available
information of research interests costs researchers a
lot of time and efforts. Although, several database
collections are currently available, such as the
Pathguide (Bader, 2006) and NAR Database
collection (Fernández-Suárez, 2013). But, it is still a
big effort for researchers to access the databases in a
systemic efficient way. Due to this need, we have
established BioMetaDB for users to systematically
reach and explore all the biomedical databases of
their interests. We firstly generated an ontology list
for individual biological databases, using ontological
parent-child concept heuristics. Then we use three
schemes to populate BioMetaDB sections: (i)
classifying the databases according to the biological
species and biological event issues; (ii) analysing the
relevance scores and grouping the databases
according to the scores and results of (i); (iii)
presenting the category with BioMetaDB linkages.
The BioMetaDB metadata can be used to find and
manage massive-scale content stored and shared on
the many biomedical repositories so that researchers
can always manage to find the information they are
looking for.
2 MATERIALS AND METHODS
BioMetaDB mined and integrated more than 1500
database sources from PubMed and public
databases. The types of databases included DNA
database, RNA database, protein database, structure
database, microarray database, etc. The approach
and workflow of our ontology-based classification
of biomedical databases are described as follows.
2.1 Data Source and Ontology
Repository
Based on the databases’ context, we had established
156
Chang C., Lin C. and Chang C..
BioMetaDB: Ontology-based Classification and Extension of Biodatabases.
DOI: 10.5220/0004801001560163
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2014), pages 156-163
ISBN: 978-989-758-012-3
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

a list of ontology features for each database.
Ontologies are domain knowledge, which can
provide a single identifier for describing each
concept or entity in a domain, even more, connect
concepts with related meanings, therefore, ontology
utility can drive data annotation and data integration.
Some databases had adapted the ontology concept
and provided access to a library of biomedical
ontologies and terminologies. For examples, the
Gene Ontology database (The Gene Ontology
Consortium; Ashburner, 2000), BRENDA
(Schomburg, 2004), TAIR (The Arabidopsis
Information Resource; Swarbreck, 2008), the
NCBI’s
BioPortal (Musen, 2012), etc. The ontology of
databases can be described as the relational schema
of their tagged corpuses. In order to make the
classification of mined databases in our BioMetaDB,
we constructed our own ontology list in which
specification and conceptualization define the
ontology purpose and provide the vocabulary,
relationships, and concepts for ontology design. The
ontological hierarchies and child-parent
relationships (PART_OF/IS_A) were established to
develop the domain ontology and sub-ontologies for
further use in implementation. Except the database
content, we also inferred the ontologies from other
groups such as the Gene Ontology database,
BioPortal, the Open Biological and Biomedical
Ontologies, the Proteomics Standards Initiative
(Orchard, 2003), and the Consultative Group on
International Agricultural Research. The relevance
among the databases was calculated according to
their ontology features, and the databases were then
grouped into various categories. In our BioMetaDB,
the species is indicated with the standard NCBI
taxonomy database taxid. In order to support search
in large, open and heterogeneous repositories of
unstructured biomedical information, we needed to
not only exploit deep levels of conceptualization of
these databases, but also their corresponding
publications and web site contents.
2.2 Relevance Measurement for
Classification of Databases
We adapted the hierarchical classification and
relevance measurement to categorize the databases.
Firstly, we had indexed the database by their
features, which were further used to evaluate the
relevance between different databases. The feature
index of each database also helped us to classify the
database. For example, the databases A, B and C can
be indexed as {human, transcription factor,
sequence}, {yeast, transcription factor}, and
{human, transcription factor binding site}
respectively. The databases A and C belong to the
“human” category, and the database B belongs to the
“yeast” category. Once the users propose the query
as “human”, they will obtain the results as databases
A and C. If the query is “human” plus “transcription
factor”, the output will be the database A. The goal
of the present work is to determinate the relevance
between each database pairs where each database
contains multiple biological features, for example,
the study species and the focused biological issue. In
the bag of indexes vector of each database, the
database was represented by vector in N-
dimensional space where N represented the total
number of feature indexes. For the relevance
calculation, we had inferred the previous database
classification method (Wu, 2005). Once two
databases share a significant number of feature
items, they were relevant to each other. For example,
we extracted the feature items of individual
database, such as A, B, C. Three databases were
presented as follows: D1= {A, B, C} D2= {A, C,
D, E}. The similarity S between the items of two
databases can be defined as,
(Item (D1) ∩Item (D2))⁄ (Item (D1) ∪Item (D2)) = S
Thus the relevance among various biomedical
databases can be measured. The significance of the S
value presents the high relevance between databases.
2.3 Database and Query
Implementation
We present an ontology-based multi database
classification and extension. The BioMetaDB is
curated by the authors and regularly updated (Fig.
1). Figure 1 is the workflow of our BioMetaDB
establishment. Generation of web pages was
implemented using the PHP server-side scripting
language for obtaining data and maintaining sessions
between web pages. The MySQL relational database
management system was used for storing the
biodatabase information in a structured manner.
BioMetaDB (http://cbs.ym.edu.tw/services/BMdb/)
provides versatile search functions with multi-source
multi-category searching through ontologies and
through researchers’ own keywords. Searching is
possible in the Web and dedicated collections, and
query results can be retrieved. A range of ontologies
can be used without assuming annotation of
databases. BioMetaDB offers a databases analysis
function through online query biased summarization
of individual databases and category sets. The
summarization criteria can be flexibly changed.
BioMetaDB:Ontology-basedClassificationandExtensionofBiodatabases
157

Further analysis is possible through clustering of
databases and identification of category ontology
concepts which can be used in query modification.
Also ontology-based cross-corpus classification, and
markup tag management are available. Classification
and clustering can be used to automatically feed into
query reformulations, and summaries manually.
These functions effectively support database
exploration. Future work will provide maps and
timelines to study the geographic and temporal
development and distribution of the collected
databases.
3 RESULTS
3.1 Index List of Database Features
An accurate analysis and classification of biological
data repositories is necessary to facilitate the access
and exploration of molecular biology data. However,
classifying biomedical databases is a difficult and
challenging task, especially when a large number of
biomedical databases are cross-related and can
involve in many diverse research interests. For an
effective navigation and selective database data
integration, we had collected information on over
1500 published online biology database in the
BioMetaDB. According to the databases’ purpose
and content, we made the feature list of each
database (Table 1). For examples, the feature list of
IUPHAR-DB (Sharman, 2013) was: {human, rat,
mouse, nonsensory G protein-coupled receptors
(GPCRs), genes and functions of nonsensory G
protein-coupled receptors (GPCRs), ligand-gated ion
channel, genes and functions of ligand-gated ion
channel subunits, genes and functions voltage-gated-
like ion channel subunits}; the feature list of
NetworKIN database (Linding, 2008) was: {human;
protein kinase; substrate of protein kinase; the
network of protein kinase}. Our approach is
different from the previous works, where we explore
the use of hierarchical ontology concept structure for
searching and identifying the probable categories in
order to classify biomedical databases. To realize
our propose method, we used the features that
extracted from the datasets, their corresponding
publications and web contents to index the
biomedical database text. These features were used
to represent our meta-databases in order to improve
the accuracy of classification performance and also
the result of searching relevant databases.
3.2 Database Category Type
Databases in the list are grouped into 15 major
categories based on the types of the biological focus
data made available (Table 2). The category of a
database can be in multiple categories if it contains
multiple data types or organism species. For
instance, the HemaExplorer database (Bagger,
2013), a curated database of processed mRNA Gene
expression profiles (GEPs) haematopoietic cells,
include data from human and mouse hematopoietic
systems, normal human samples, and human acute
myeloid leukemia (AML). Therefore, HemaExplorer
database will be found in at least three categories,
human, mouse and disease data. For our approach to
categorization, we devised an expert-tagged corpus
of keywords and phrases associated with biomedical
data categories (the category names, variations such
as plurals, synonyms, and related words obtained by
examining the original publications of each collected
databases). Due to the fact that some places where
keywords appeared are more important than those in
other sections, the web sites of each database used
were also searched for marking up relevant tag
annotations and matches (see Figure 2 for the
distribution of classified biomedical databases). In
our research, each database must be represented by a
set of feature tags. The Table 1 shows an example
for the index list of BioMetaDB.
3.3 Database Resource Extensions
Each category keyword match in a database
publication and its web site was later treated as votes
for the relevant category for each category
associated with the identified cases. These votes
were compiled for every database, which was
labelled with high-scoring categories. These efforts
together lay the groundwork for our deep semantic
meta-mining that is driven by both meta-data and the
collective expertise of data miners embodied in the
data mining ontology and knowledge base. Even
though conceptual representations are difficult and
effort-intensive to create and maintain, our
BioMetaDB web site hosts the new search facility,
provides the database category search function and
links to the homepages of all the collected databases.
BioMetaDB can reduce the distance between the
logic representation of the available database
systems and the real one in the user’s mind with
regards to the formulation of queries and the
understanding of database contents.
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
158

3.4 Web Services
To implement useful web services of BioMetaDB
that are accessed through internet connection, we
had set up a web server. The BioMetaDB database
was designed to store metadata and relationships
between collected databases (Figure 3). Fields
containing multiple records were stored as delimited
text within the same record to reduce complexity
and improve efficiency of queries. The homepage of
BioMetaDB is shown as in Figure 4. All the
metadata records of BioMetaDB as well as the
relationships between them are parsed and stored in
a local database. An interactive web search interface
with convenient utilities provides query capabilities
not available via other tools and makes querying the
BioMetaDB metadata both easier and more
powerful. The ‘Data Browsing’ Web service
includes functionality to get a list of all the databases
that allow users to access interconnected biological
and biomedical databases of their interests using the
ontology hierarchy. We also outline how the
databases of BioMetaDB can be extracted to address
point-by-point the dependent queries put forth in the
Introduction section. The Data Query web service is
designed to extract branches of ontologies given a
term to serve as the root node in the ontology view.
This web service is very popular for generating
views of content specific portions of large ontologies
such as the NCBI Taxonomy and the Systematized
Nomenclature of Medicine-Clinical Terms
(SNOMED-CT). The database names in BioMetaDB
are linked to the database home-page and clicking
on ’more’ next to each database leads to a
description of the database listing full name, short
name, homepage Uniform Resource Locator (URL),
and text description.
4 DISCUSSION AND
CONCLUSIONS
Several distinguished respects of BioMetaDB make
it unique than other existing database collections.
Firstly, it collects abundant databases with a wide
range. Secondly, the ontology extraction and utility
of the database make the category of those collected
databases clear and easy to be queried. Thirdly,
users can combine several items to query the desired
database from the database discipline, for examples,
the species, the nucleotide, etc. The current content
(updated November 2013) of Pathguide has
information of 547 biological pathway-related
resources and molecular interaction-related
resources. Pathguide majorly focuses on the cellular
pathways and network databases. In contrast, our
BioMetaDB includes almost all the available
biomedical databases. Nucleic Acid Research
(NAR) database collection has collected biological
databases for more than a decade. Therefore, it has
abundant database information. However, the NAR
Database Summary offers only three methods for
searching the database, the Alphabetic List, the
Category List, and the Search Summary Papers by
given a Search Term. The disadvantages of this kind
of database search methods include the time wasting
and the requirement of prior knowledge of the
database names. As for our BioMetaDB, users can
find their interesting database through combinatorial
key words search to access the databases.
Biometadata services are needed to support the
intensive applications of biomedical resources. The
large number of biomedical databases that published
and/or on the internet makes the process of
classification become challenging and laborious.
The reason is that there are many categories of
biomedical databases available and each category
have many different classes. There have been prior
researches on classification of biological databases.
The typical method is to group them on simple
macromolecule types without considering the
semantic relationship among databases. However,
this simplified concept model is not very efficient,
especially for biomedical data across multiple
research topics. Here, we present BioMetaDB, an
integrated resource for researchers to systematically
locate and access the current avalanche of biological
and medicine databases based on semantically
annotated corpus for marking up the instances of
biomedical ontology. Our future aim is to seek and
add more features for selecting relevant and
meaningful tags in order to enrich or expand the
connection of biomedical databases. These features
would be used to index the biomedical databases for
increasing the accuracy of classification
performance and also the result of searching relevant
databases.
ACKNOWLEDGEMENTS
This work was supported by grants from the
National Science Council (NSC) of Taiwan [NSC
102-2319-B-010-002, NSC 102-EPA-F-006-002],
and the Aim for the Top University Plan grant from
the Ministry of Education, Taiwan.
BioMetaDB:Ontology-basedClassificationandExtensionofBiodatabases
159

Table 1: An example of BioMetaDB index list.
For an effective navigation and selective database
data integration, the feature list of every collected
database was made according to the purpose and
content of each database.
Database name Website Feature index
OGRe - Organellar Genome Retrieval http://ogre.mcmaster.ca/ Animal, mitochondria, mitochondrial genome
MitoDrome http://www-lecb.ncifcrf.gov/mitoDat/ D. melanogaster, mitochondria, nuclear genes encoding mitochondrial proteins
HMPD http ://b io info . nist. gov/hmp d / Human, mitochondria, mitochondria l p rotein
HmtDB -
Human Mitochond rial DataBase
http://www.hmtdb.uniba.it/ Human, mitochondria, mitochondria bioinformatic resource
Human MtDB http://www.genpat.uu.se/mtDB/ Human, mitochondria, mitochondrial genome
HvrBase++ http://www.hvrbase.org/ Human, great apes, neanderthaler, mitochondria, mitochondrial DNA sequence, sequences of hypervariable regio
MitoDat http://www-lecb.ncifcrf.gov/mitoDat/ Human, animal, yeast, fungi, plant, mitochondria, mitochondria DNA, mitochondria protein
MamMiBase http://www.mammibase.lncc.br/ Mammalian, mitochondria, mitochondrial genome
Mitome http://mitome.knu.ac.kr// Metazoan, mitochondria, mitochondrial genomes, mitochondrial gene arrangements
MitoZoa http://mi.caspur.it/mitozoa/
Metazoan, mitochondria, mitochondrial genomes, mitochondrial gene order, non- coding regions, gene content,
and gene sequences
MitoRes http://mitores.ba.itb.cnr.it/index.php Metazoa, mitochondrial, nuclear-encoded mitochondrial proteins, gene, transcript
MitoGenesisDB http://www.dsimb.inserm.fr/dsimb_tools/mitgene/ Mitochondria, Expression data of spatio-temporal dynamics of mitocho ndrial bioge nesis
MITOMAP http://www.mitomap.org/
Human, mitochondria,Human mitochondrial genome; human mitochondrial DNA variation;
mitochondria DNA rearrangements
MitoProteome http://www.mitoproteome.org/
Human, mitochondria, mitochondrial protein sequence, mitochondrial protein sequence encoded
by mitochondrial and nuclear genes
MitoMiner http://mitominer.mrc-mbu.cam.ac.uk/
Human, mouse, Droso p hila, N . crassa, P. falciparum; G. lamblia; rat, yeast, Arabidopsis; T. thermophila ,
mitochondria, mitochondrial proteomics
MITOP2 http://www.mitop2.de/ Yeast, mouse, human, mitochondria, mitochondrial protein, mitochondrial dysfunction
MPIM -
Mitochondrial Protein Import Machinery
http://www.plantenergy.uwa.edu.au/applications/
mpimp/index. html
Yeast, Human, Rat, Mouse, Drosophila, Danio rerio, Cenorhabtidis elegans, Arabidopsis,
Rice, Plasmodium falciparum, mitochondria,information on the mitochondrial protein import apparatus
YDPM - Yeast Deletion Project
http://www-deletion.stanford.edu/YDPM/
YDPM_index.html
Yeast, mitochondria proteomics
YMPD http://bmerc-www.bu.edu/projects/mito/ Yeast, mitochondria, mitochondrial protein
FUGOID
http://fugoid.webhost.utexas.edu/introndata/
main.htm
Mitochondria, chloroplast, functional and structural information of mitochondria intron,
functional and structural information of chloroplast intron
GOBASE http://gobase.bcm.umontreal.ca/ Mitochondria, chloroplast, mitochondrial sequence, chloroplast sequence, gene, introns, proteins, genetic map
Organelle genomes
http://www.ncbi.nlm.nih.gov/genomes/
ORGANELLES/organelles.html
Mitochondria, Plastids, mitochondria genome, plastid genome
Chloroplast Genome Database http://chloroplast.cbio.psu.edu/ Chloroplast, Chloroplast genome data
CpBase: The Chloroplast Genome Database http://chloroplast.ocean.washington.edu/ Chloroplast, chloroploast genome, gene
PLprot http://www.plprot.ethz.ch/ Arabidopsis thaliana, chloroplast, chloroplast protein
PeroxisomeDB http://www.peroxisomedb.org/ Human, Saccharomyces cerevisiae, peroxisome, peroxisomal proteome
Organelle DB http ://o rganelledb.lsi.umich. e d u/
S. cerevisiae, A. thaliana, D. melanogaster, C. elegans, and M. musculus,
endoplasmic reticulum, membrane protein, miscellaneous others, mitochondrion, nucleus, protein complex
Plant Organelles Database http://podb.nibb.ac.jp/Organellome/
Plant, organell, tissue, developmetal stage, images and movie of of plant organelles during developmetal stage,
image of plant tissue during development stage,
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
160
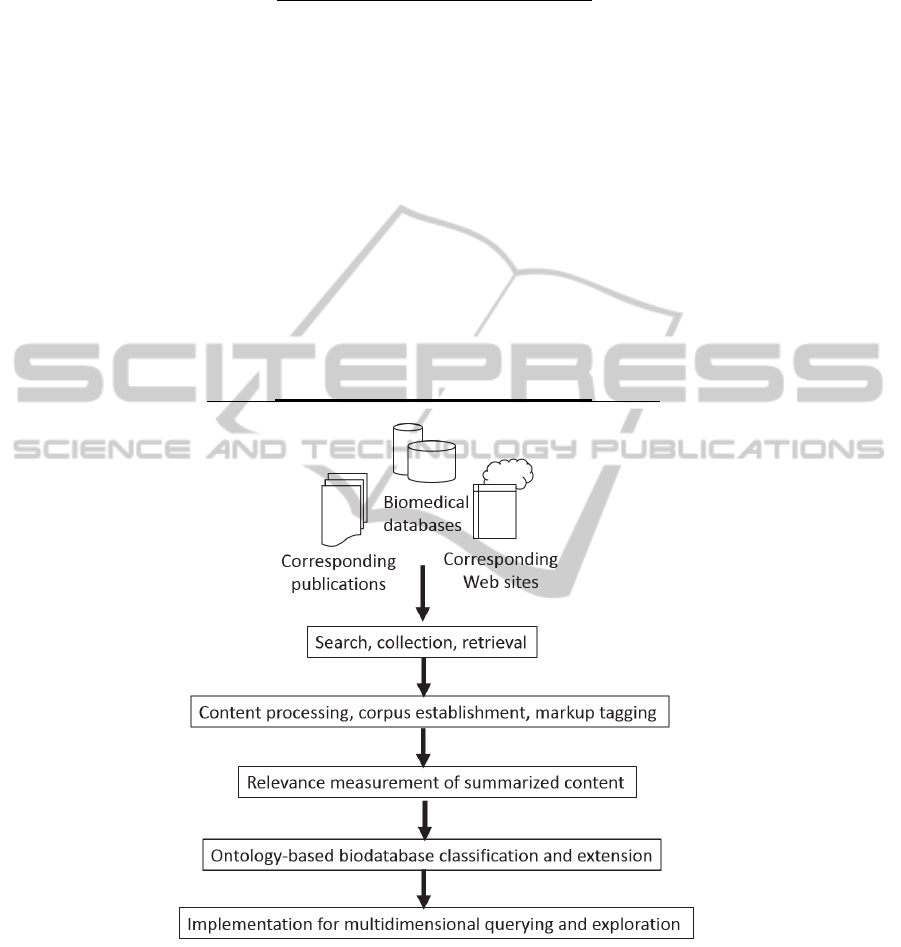
Table 2: The BioMetaDB category classification.
Bio-ontology Category
Organism species
DNA data
RNA data
Protein data
Enzyme
Structure data
Genome
Metabolism
Signal transduction
Organelle
Microarray data
Disease data
Drug and pharmacogenomics
Immune data
Molecular biology tools
Figure 1: Workflow for ontology-based multi-database classification and extension.
The collected databases were grouped into 15
major categories based on the types of the biological
focus data made available. The category of a
database can be in multiple categories if it contains
multiple data types or organism species.
All the retrieved information contents of
collected databases were processed to establish
relevance measurement. Ontology-based
classification and extended higher level connections
were used to implement multidimensional querying
and exploration.
In our research, each database must be
represented by a set of feature tags. A histogram plot
of the distribution of biomedical databases after
classification shows none uniformly grouped
categories.
BioMetaDB:Ontology-basedClassificationandExtensionofBiodatabases
161

Figure 2: The distribution of biomedical databases after classification.
Figure 3: Diagram of entity relationships in BioMetaDB.
0
50
100
150
200
250
300
350
400
450
500
Number
Thedistributionofdatabase
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
162

An entity-relationship diagram with relevant
attributes and value types shows the connections
between major entity types.
Figure 4: BioMetaDB web page.
All the metadata records of BioMetaDB as well as
the relationships between them were parsed and
stored in a local database. An interactive web search
interface with convenient utilities provides query
capabilities.
REFERENCES
The Gene Ontology Consortium. 2000. Gene ontology:
tool for the unification of biology. Nat. Genet.
25(1):25-9. http://www.geneontology.org/
Schomburg, I., Chang, A., Ebeling, C., Gremse, M., Heldt,
C., Huhn, G., and Schomburg D., 2004. BRENDA, the
enzyme database: updates and major new
developments. Nucleic Acids Res. 32: D431-433.
http://www.brenda-enzymes.org/
Swarbreck D., Wilks C., Lamesch P., Berardini T. Z.,
Garcia-Hernandez M., Foerster H., Li D., Meyer T.,
Muller R., Ploetz L., Radenbaugh A., Singh S., Swing
V., Tissier C., Zhang P., Huala E., 2008. The
Arabidopsis Information Resource (TAIR): gene
structure and function annotation. Nucleic Acids Res.
36:D1009-14.
http://arabidopsis.org.
Musen M. A., Noy N. F., Shah N. H., Whetzel P. L.,
Chute C.G., Story M.A., Smith B.; NCBO team. 2012.
The National Center for Biomedical Ontology. J Am
Med Inform Assoc. 19(2): 190-5. http://
www.bioontology.org/
The Open Biological and Biomedical Ontologies.
http://obofoundry.org/
Orchard S., Hermjakob H., Apweiler R., 2003. The
proteomics standards initiative. Proteomics.
3(7):1374-6. http://www.psidev.info/
The Consultative Group on International Agricultural
Research. http://www.cgiar.org/
NCBI taxonomy database. http://www.ncbi.nlm.nih.gov/
taxonomy.
Bader GD., Cary M. P., Sander C., 2006. Pathguide: a
Pathway Resource List. Nucleic Acids Res. 34:
D504–D506.
Fernández-Suárez X. M., Galperin M. Y., 2013. The 2013
Nucleic Acids Research Database Issue and the online
molecular biology database collection. Nucleic Acids
Res. 41: D1-7.
Wu X., Zhang C., Zhang S., 2005. Database classification
for multi-database mining. Information Systems. 30:
71–88.
Sharman J. L., Benson H. E., Pawson A. J., Lukito V.,
Mpamhanga C. P., Bombail V., Davenport A. P.,
Peters J. A., Spedding M., Harmar A. J.; NC-
IUPHAR. 2013. IUPHAR-DB: updated database
content and new features. Nucleic Acids Res.41:
D1083-8. http://www.iuphar-db.org/
Linding R., Jensen L. J., Pasculescu A., Olhovsky M.,
Colwill K., Bork P., Yaffe M. B., Pawson T., 2008.
NetworKIN: a resource for exploring cellular
phosphorylation networks. Nucleic Acids Res. 36:
D695-9.
http://networkin.info/
Bagger F. O., Rapin N., Theilgaard-Mönch K.,
Kaczkowski B., Thoren L. A., Jendholm J., Winther
O., Porse B.T., 2013. HemaExplorer: a database of
mRNA expression profiles in normal and malignant
haematopoiesis. Nucleic Acids Res. 41:D1034-9.
http://servers.binf.ku.dk/hemaexplorer.
The Systematized Nomenclature of Medicine-Clinical
Terms (SNOMED-CT). http://www.ihtsdo.org/
snomed-ct.
BioMetaDB:Ontology-basedClassificationandExtensionofBiodatabases
163
