
Methods for Quality Control of Low-resolution MALDI-ToF Spectra
Michal Marczyk and Joanna Polanska
Data Mining Group, Faculty of Automatic Control, Electronics and Computer Science, Silesian University of Techonology,
Akademicka 16, Gliwice, Poland
Keywords:
MALDI-ToF, Protein Profiling, Quality Control.
Abstract:
Protein profiling of human blood serum or plasma using MALDI-ToF mass spectrometry may be used for
identification of candidates for disease biomarkers. Due to many biological and technical difficulties emerging
during preparation of the sample and spectra measurement quality control step is becoming important. In
this study we compared different methods for finding low quality spectra based on the Pearson correlation
coefficient and proposed two novel solutions. First one utilizes information about area under the measured
spectrum and other incorporates modeling of signal-to-noise ratio of spectra intensity by mixture of Gaussians.
Obtained results show that removing of outlying samples increases the similarity of spectra obtained within
the same experimental conditions. What is more important it increases reproducibility of peak detection by
decreasing the coefficient of variation of peaks intensities within a group and increasing its prevalence. This
work shows that appropriate identification and removing of low quality spectra is a necessary step in analysis
of mass spectrometry data and proposed tools are appropriate for quality control of MALDI-ToF data.
1 INTRODUCTION
Matrix-assisted laser desorption and ionization time
of flight (MALDI-ToF) mass spectrometry (MS) can
detect components at a very low concentration lev-
els, which offers opportunities to discover diagnos-
tic markers for a number of major diseases includ-
ing cancer. Human blood serum or plasma may cap-
ture proteins and their fragments released from all
organs and tissues in different conditions. Investi-
gating of serum/plasma proteome using MALDI-ToF
MS called proteome profiling can be used for iden-
tification of panels of marker candidates (Palmblad
et al., 2009). Due to requirements of low cost and
high speed of measurements obtained spectra are low-
resolution data and do not contain the isotopic enve-
lope of peaks.
Highly automated acquisition of a large number of
data increases the risk of receiving poor quality sig-
nals. MALDI-TOF results may also be weakened by
inadequate deposition of the sample on the plate, poor
cleaning of the plate between runs and other technical
factors. Quality control (QC) is a first important step
in data analysis. Detection of the spectra with low
signal-to-noise ratio (SNR) allows eliminating heav-
ily noisy data from further analysis preventing false
discoveries of candidate markers which can lack bio-
logical relevance. It is important to properly exclude
outlying spectra to guarantee the reliability of discov-
ered patterns and reproducibility of MALDI-ToF ex-
periments.
The easiest and fastest way of quality control is the
visual inspection of measurements heat maps. Dif-
ferent colors are used to depict the intensity of indi-
vidual spectra, which are arranged in successive rows
on the graph (Figure 1). In the proteome profil-
ing using human specimens most major peaks should
be visible on all spectra, so it is possible to visu-
ally determine outliers. This method is also effec-
tive to check the quality of calibration of the mass
spectrometer. Another way of QC is to use the area
under the curve of the whole spectrum (total ion cur-
rent, TIC). Number of ions that reach the detector at
the same experimental conditions and with use of the
same laser power should be similar. It is assumed
that only small deviations in the values of TIC for
the spectra obtained in one experiment should be ob-
served. A useful approach to check data similarity
is to calculate the Pearson correlation coefficients be-
tween the raw spectra, and then plot a correlation ma-
trix (Hong et al., 2005) or use the diagnostic plots
(Whistler et al., 2007) to find spectra which measure-
ments are different from the others. Another group
of methods involves placement of control signals in
analyzed sample. Such information may be used to
check only existence of control peaks or in more so-
172
Marczyk M. and Polanska J..
Methods for Quality Control of Low-resolution MALDI-ToF Spectra.
DOI: 10.5220/0004804201720177
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2014), pages 172-177
ISBN: 978-989-758-012-3
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

phisticated methods like principal component analy-
sis models. It was proved that using statistical meth-
ods on control peaks is more reliable and suitable for a
large number of spectra (Coombes et al., 2003). Since
most human cells, despite the variety of functions in
the body, are very similar to each other in terms of
its structure, it is possible to define a standard set of
proteins that are present in most experimental condi-
tions. In this way we can define a positive control
for start-up of the mass spectra, which may indicate
poor performance of real peak detection (Slany et al.,
2009).
Figure 1: Heat map for 3 cancers dataset.
In this study we investigated methods for finding
the low quality MALDI-ToF spectra which can be
used directly on the measured signals as a first step
of mass spectra analysis pipeline. We chose two pro-
cedures from the literature that are using the Pear-
son correlation coefficient and introduced two new
algorithms: one using an outlier detection algorithm
on TIC and one using a Gaussian mixture modeling
(GMM) on a robust version of SNR. We analyzed the
effect of using different quality control algorithms on
the integrity of the spectra within a group and its in-
fluence on performance of peak detection.
2 MATERIALS AND METHODS
2.1 Data
Three different MALDI-ToF datasets obtained for
proteomic profiling were used to compare quality
control algorithms: 3 cancers dataset, larynx dataset
and ovarian dataset. First dataset consists of 120 can-
cer patients from which 35 patients are with squa-
mous cell cancer located in head and neck region
(HNC), 35 patient are with colorectal cancer (CRC)
and 50 patients are with non-small cell lung cancer
(LC) and 45 volunteers in control group. Samples
were collected before the start of a therapy of patients
and measured using 4 technical replicates of the same
sample giving 659 spectra (1 damaged file). Spec-
tra were recorded in the mass range between 2 and 13
kDa (Pietrowska et al., 2012). Second dataset consists
of 54 patients with a squamous cell cancer located at
larynx. Blood samples were collected before the start
of therapy (sample A), 2 weeks after the start of ra-
diotherapy (sample B) and 46 weeks after the end of
radiotherapy (sample C). Not all patients had taken
samples B and C. There were 4 technical replicates
of each sample giving 620 spectra in total. Spectra
were recorded in the mass range between 2 and 13
kDa (Widlak et al., 2011). Third dataset consists of
93 ovarian cancer patients and 77 normal blood serum
samples. Spectra were recorded in the mass range be-
tween 3.5 and 20 kDa (Wu et al., 2006).
3 cancers and larynx datasets were kindly pro-
vided by the authors of the original research
articles (Pietrowska et al., 2012)(Widlak et al.,
2011) only for testing different QC algorithms.
Ovarian cancer dataset was downloaded from
http://bioinformatics.med.yale.edu/MSDATA2.
2.2 Spectra Pre-processing
Mass spectrum pre-processing involves removal of a
measurement error, while retaining the essential bio-
logical information contained in the sample. It is an
obligatory step to appropriately extract signal peaks
which describe composition of analyzed material (Hi-
lario et al., 2006). First, spectra are resampled to
a common M/Z interval. Next, in baseline correc-
tion step we estimate the noise within multiple shifted
window and regresses the varying baseline to the win-
dow points using a spline approximation. Spectra are
aligned using peak alignment by fast Fourier trans-
form algorithm (Wong et al., 2005) modified by in-
troducing values of parameters to be relative to mean
M/Z of analyzed segment. To detect signal peaks
we used algorithm based on transforming signal into
wavelet space from MassSpecWavelet package (Du
et al., 2006).
2.3 Algorithms for QC
In (Hong et al., 2005) a correlation matrix (Figure
2) was developed as a QC tool in surface-enhanced
laser desorption/ionization (SELDI). It is a mass spec-
trometry method which differs to MALDI-ToF only
in ionization phase, but measured signals have simi-
lar shapes and properties. The rationale behind this
approach is to assume that the protein expression pro-
files of samples obtained in the same experimental
MethodsforQualityControlofLow-resolutionMALDI-ToFSpectra
173

Table 1: Comparison of different quality control methods using a percent of deleted spectra and the median similarity in three
datasets. CRC - colorectal cancer, CTR - control group, HNC - head and neck cancer, LC - lung cancer, A,B,C - groups
representing different time points, CTR - control group, OVC - ovarian cancer. None - results without quality control, Corr -
method based on a correlation matrix, DP - method using the diagnostic plot, TIC - method based on a total ion current, SNR
- method based on a signal-to-noise ratio of spectrum intensities.
3 cancers Larynx Ovarian
Measure Method CRC CTR HNC LC A B C CTR OVC
Deleted Corr 3.57 3.33 7.86 16.08 0.00 1.89 3.13 2.60 1.08
spectra DP 3.57 3.33 7.86 16.08 0.00 0.00 2.08 2.60 1.08
[%] TIC 3.57 3.89 7.86 16.08 0.46 0.94 0.52 7.79 4.30
SNR 3,57 6.11 7.86 17.09 8.33 8.96 5.21 38.96 25.81
Median none 0.653 0.765 0.576 0.564 0.779 0.781 0.803 0.613 0.647
similarity Corr 0.680 0.785 0.650 0.695 0.779 0.791 0.815 0.632 0.652
DP 0,680 0.785 0.650 0.695 0.779 0.781 0.811 0.632 0.652
TIC 0,680 0.788 0.650 0.695 0.778 0.781 0.804 0.662 0.640
SNR 0,680 0.801 0.650 0.697 0.777 0.780 0.804 0.757 0.695
conditions should be comparable and thus that the
correlation among spectra intensities should be high.
Authors of algorithm suggest that the inter-spectral
correlation coefficient values of R from 0.95 to 0.97
are attainable and representative figure-of-merit for
quality data in analysis of SELDI sample replicates.
Due to the complexity of protein signals from human
specimens and individual variability of the patients,
we set new thresholds for the correlation coefficients
using clustering procedure (Figure 3). Hierarchical
clustering was performed on the mean correlation co-
efficients of the spectra intensities using a shortest Eu-
clidean distance metric. Application of other distance
metrics in most cases give similar results. Optimal
number of clusters was found by maximizing a sil-
houette measure. Sample with mean correlation co-
efficient belonging to the most frequent cluster are
treated are high quality spectra. The remaining spec-
tra are suspected for poor quality and removed from
further analysis.
Figure 2: A correlation matrix for larynx dataset.
Another application of the idea of measuring a re-
lationship between spectra intensities within a group
is creating the diagnostic plots (Figure 4). Algo-
0.2 0.3 0.4 0.5 0.6 0.7 0.8
0
20
40
60
80
100
120
140
160
Mean Pearson correlation coeff.
Counts
10/620 spectra removed
Figure 3: Histogram of the mean Pearson correlation coeffi-
cient for larynx dataset with threshold value marked by the
dashed line.
rithm of (Whistler et al., 2007) starts with generating
a pairwise similarity matrix using the Pearson corre-
lation coefficient on normalized intensity values for
each spectrum. To visually depict the data, the diag-
nostic plot is drawn as 1 minus the mean of Pearson
correlation coefficients against the range of the corre-
lation coefficients. In our implementation due to high
non-normality of distribution of the correlation coef-
ficient we introduced robust measures of location and
dispersion such as the median value and the interquar-
tile range instead of the mean and a range. Authors
of the algorithm established values of cut-off for 1-
the mean of correlation coefficient by comparing the
results depicted in the diagnostic plots to other eval-
uation methodologies, such as principal component
analysis of the processed spectra or SNR, and by com-
paring the number of peaks in each spectrum with the
average number of peaks for all spectra in the dataset.
In this paper to find the cut-off value we introduced
a method based on a hierarchical clustering described
in a previous paragraph.
The number of measured ions for spectra in the
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
174
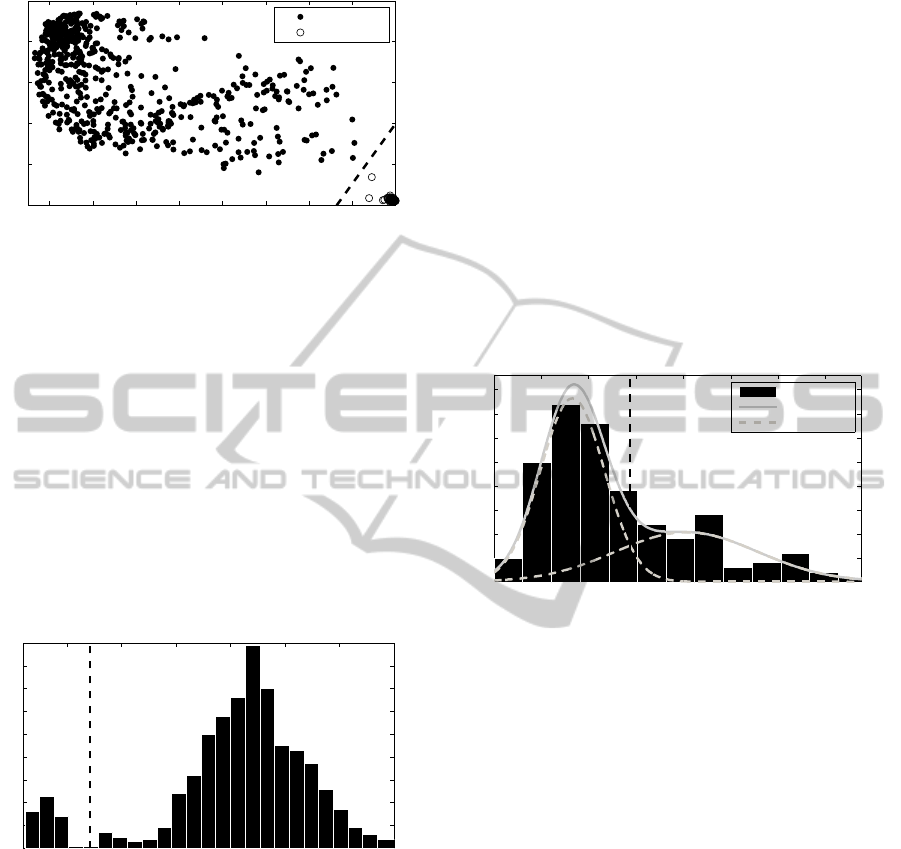
0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
0
0.1
0.2
0.3
0.4
0.5
1−median(r)
IQR(r)
54/659 spectra removed
Proper
Low quality
Figure 4: Diagnostic plot for 3 cancers dataset with thresh-
old value marked by the dashed line.
same experiment should be comparable, so any mean-
ingful deviances in area under the spectrum intensi-
ties should be treated as outlying measurements. Our
studies showed that spectra with lower TIC have a
negative impact on results of further analysis than the
one with too high TIC due to the presence of multi-
ple peaks resulting from noise (data not presented).
To reduce skewness of distribution of TIC and ex-
tract samples with low values of TIC we initially
perform logarithmic transform of TIC. Spectra with
lower area under the signal are found using outlier de-
tection method for skewed data (Hubert and Van der
Veeken, 2008) on logarithm of TIC (Figure 5).
−1 0 1 2 3 4 5
0
10
20
30
40
50
60
70
80
90
Logaritm of Total Ion Current
Counts
55/659 spectra removed
Figure 5: Histogram of TIC in logarithmic scale for 3 can-
cers dataset with threshold value marked by the dashed line.
SNR is a commonly used measure of the signal
quality. It makes use of the mean and the standard
deviation of intensities computed for each sample.
These measures of location and dispersion are very
sensitive to outliers. For each spectrum we have a se-
ries of tens of thousands points, only some of which
are associated with significantly higher intensity val-
ues forming signal peaks. We propose to use a robust
SNR measure defined as the median signal intensity
to its median absolute deviation. In contrast to the
classical approach of filtering data with low SNR, in
our application we are looking for the samples with
too high SNR values. High value of robust SNR cal-
culated for mass spectrum intensities indicates a large
amount of only very low peaks, which may be the re-
sult of noise occurrence. To identify the cut-off value
we introduced an idea of modeling the distribution of
SNR using Gaussian mixture model (Figure 6). We
find the optimal number of components by minimiz-
ing Bayesian information criterion. In order to dis-
tinguish Gaussian components consisting low quality
spectra we use a k-means clustering procedure, which
classifies estimated components into two groups in
three dimensional space with coordinates given by
means, standard deviations and weights of Gaussian
components. Then we remove samples which belong
to cluster of the components located at the right hand
side of the robust SNR scale (Marczyk et al., 2013).
20 40 60 80 100 120 140 160
0
5
10
15
20
25
30
35
40
Counts
SNR_robust [%]
54/170 spectra removed
Data
Model
Components
Figure 6: Histogram of robust SNR for ovarian dataset with
threshold value marked by the dashed line.
3 RESULTS
Four selected quality control algorithms have been ap-
plied to study three MALDI-ToF datasets containing
different number of groups and different number of
spectra within a group. All procedures were applied
to raw data without information of the group mem-
bership. Influence of introducing quality control as a
pre-processing step on the integrity of the spectra and
a performance of peak detection was obtained for data
within the groups.
In Table 1 we presented percentages of deleted
spectra after using different quality control tools.
In some cases all algorithms remove the same low
quality signals which results in the same further re-
sults (group CRC and HNC in 3 cancers dataset).
For group LC all methods indicated that about 17%
of data should be removed. In larynx and ovarian
datasets SNR method cleaned much more spectra than
other algorithms. For almost all calculations methods
Corr and DP removed the same amount of data.
To quantify influence of quality control algorithms
MethodsforQualityControlofLow-resolutionMALDI-ToFSpectra
175

Table 2: Comparison of different quality control methods using the median CV and a prevalence of peaks in three datasets.
CRC - colorectal cancer, CTR - control group, HNC - head and neck cancer, LC - lung cancer, A,B,C - groups representing
different time points, CTR - control group, OVC - ovarian cancer. None - results without quality control, Corr - method based
on a correlation matrix, DP - method using the diagnostic plot, TIC - method based on a total ion current, SNR - method based
on a signal-to-noise ratio of spectrum intensities.
3 cancers Larynx Ovarian
Measure Method CRC CTR HNC LC A B C CTR OVC
Median None 134.65 113.68 123.59 142.70 73.02 86.34 68.80 54.76 64.60
CV Corr 130.85 109.37 109.89 120.69 72.85 80.20 68.07 52.62 64.40
[%] DP 130.85 109.37 109.89 120.69 73.02 86.34 68.80 52.62 64.40
TIC 130.85 107.89 109.89 120.69 72.73 85.83 68.98 49.65 64.27
SNR 130.85 103.54 109.89 119.76 69.38 82.66 68.45 54.03 68.16
Median none 37.86 40.56 40.71 35.68 31.48 34.91 33.59 42.86 40.86
prevalence Corr 38.52 40.80 42.64 38.92 30.09 33.89 32.53 42.67 41.30
[%] DP 38.52 40.80 42.64 38.92 31.48 34.91 33.24 42.67 41.30
TIC 38.52 41.62 42.64 38.92 30.70 34.76 34.03 40.85 40.45
SNR 38.52 42.01 42.64 38.79 32.58 35.49 34.34 46.81 43.48
on data integrity we calculated the similarity mea-
sure (Frank et al., 2008) using all points of spectra.
The intra-group similarity was defined as the median
of spectra similarity calculated pairwise between the
group members (Table 1). In almost all cases exe-
cuting quality control step increased intra-group sim-
ilarity. For 3 cancers and ovarian datasets the best re-
sults are obtained after using SNR method. For larynx
dataset using Corr method gave the highest similarity.
To measure the peaks reproducibility which is a
good assessment of the overall consistency of a set
we first detect peaks in a mean spectrum which is
constructed as the average of intensities of all spec-
tra in a given dataset. Next, we detected peaks for an
individual measurements and performed peak match-
ing to the reference based on M/Z distance. We set
a maximum considered value for the difference be-
tween peaks to 0.3% * M/Z. We calculated a preva-
lence and the coefficient of variation for the peaks in-
tensities and summarize it within the groups calculat-
ing the median value (Table 2). In almost all cases
use of quality control algorithms decreased CV and
increased peaks prevalence. For CTR and LC groups
of 3 cancers dataset SNR method gave the best results.
In two cases Corr method gave lower CV than others.
For ovarian dataset using TIC method we got the low-
est CV. Introducing SNR method in this dataset lead
to increased prevalence of peaks but in cost of reduced
CV, which is still similar to the case when no quality
control method is used.
4 DISCUSSION
Removing of low quality spectra has a big influence
on increasing the amount of association between sam-
ples. Providing better similarity of data within the
same experimental group may increase reliability of
protein profiling procedure. It may lead to more accu-
rate discrimination of samples between different treat-
ment groups and finding more significant biomark-
ers. Quality control also significantly increases repro-
ducibility of experiment results by decreasing vari-
ability of estimated intensities of peaks.
It is a desirable property of QC tools that crite-
rion for removing data is based on all samples in the
dataset and disregards information on measurement
group labels. Such an assumption makes this phase
independent to further data analysis like searching for
proteins that show different intensity levels between
two groups or data classification. A violation of this
condition can lead to the loss of FDR control. All
methods used in this study satisfy an assumption of
independence.
For two algorithms adapted from the literature
we introduced a new method for separation of spec-
tra with different quality using a hierarchical cluster-
ing. In original implementations these values were
set manually by the researcher. The visual inspection
of histograms of the correlation coefficients and the
diagnostic plots with thresholds estimated using our
method, together with presented results of analysis,
proved that it is a good approach to automate process
of finding cut-off values for different datasets.
Quality control process implicates reducing di-
mensionality of the dataset by rejecting the spectra
resulting from measurement errors. In most of the
applications it is proper to delete no more than 10-
15% of data. In ovarian dataset SNR method re-
moves about 30% of data which is not tolerable. This
may induce a discussion if a MALDI-ToF experiment
was conducted properly. However, applying of SNR
method significantly increased similarity of ovarian
dataset and a prevalence of peak detection, which
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
176

proves correctness of using this method. It may hap-
pen that there are too many low quality spectra in a
dataset and our method allows for their appropriate
control. But when the number of deleted spectra is
too large in point of view of the researcher we recom-
mend to use TIC method which also provided good
results.
5 CONCLUSIONS
Applying of rigorous procedures during preparation
of the sample and measurements of signal does not
guarantee that all spectra from the experiment are of
sufficient quality for further data analysis. In order to
provide that only high quality data are used a com-
prehensive quality control step is required. For this
purpose we recommend to use our method based on
Gaussian mixture modeling of robust SNR measure.
It is a fully automatic algorithm and has a potential to
adapt cut-off values for removing spectra in different
sets of MALDI-ToF data. Eliminating of the outly-
ing signals with our method increases the similarity
of samples measured within the same experimental
conditions and reproducibility of peak detection al-
gorithms.
ACKNOWLEDGEMENTS
This work was financially supported by Silesian Uni-
versity of Technology internal grant for young scien-
tists BKM/514/RAU-1/2013 (MM) and National Sci-
ence Centre grant no. UMO-2013/08/M/ST6/00924
(JP).
REFERENCES
Coombes, K. R., Fritsche, H. A., J., Clarke, C., Chen, J. N.,
Baggerly, K. A., Morris, J. S., Xiao, L. C., Hung,
M. C., and Kuerer, H. M. (2003). Quality control and
peak finding for proteomics data collected from nip-
ple aspirate fluid by surface-enhanced laser desorption
and ionization. Clinical Chemistry, 49(10):1615–23.
Du, P., Kibbe, W. A., and Lin, S. M. (2006). Improved peak
detection in mass spectrum by incorporating continu-
ous wavelet transform-based pattern matching. Bioin-
formatics, 22(17):2059–65.
Frank, A. M., Bandeira, N., Shen, Z., Tanner, S., Briggs,
S. P., Smith, R. D., and Pevzner, P. A. (2008). Cluster-
ing millions of tandem mass spectra. J Proteome Res,
7(1):113–22.
Hilario, M., Kalousis, A., Pellegrini, C., and Muller, M.
(2006). Processing and classification of protein mass
spectra. Mass Spectrometry Reviews, 25(3):409–49.
Hong, H., Dragan, Y., Epstein, J., Teitel, C., Chen,
B., Xie, Q., Fang, H., Shi, L., Perkins, R., and
Tong, W. (2005). Quality control and quality as-
sessment of data from surface-enhanced laser desorp-
tion/ionization (seldi) time-of flight (tof) mass spec-
trometry (ms). BMC Bioinformatics, 6 Suppl 2:S5.
Hubert, M. and Van der Veeken, S. (2008). Outlier detec-
tion for skewed data. Journal of Chemometrics, 22(3-
4):235–246.
Marczyk, M., Jaksik, R., Polanski, A., and Polanska, J.
(2013). Adaptive filtering of microarray gene expres-
sion data based on gaussian mixture decomposition.
BMC Bioinformatics, 14(1):101.
Palmblad, M., Tiss, A., and Cramer, R. (2009). Mass spec-
trometry in clinical proteomics - from the present to
the future. Proteomics. Clinical applications, 3(1):6–
17.
Pietrowska, M., Polanska, J., Suwinski, R., Widel, M.,
Rutkowski, T., Marczyk, M., Dominczyk, I., Ponge,
L., Marczak, L., Polanski, A., and Widlak, P. (2012).
Comparison of peptide cancer signatures identified by
mass spectrometry in serum of patients with head and
neck, lung and colorectal cancers: Association with
tumor progression. International Journal of Oncol-
ogy, 40(1):148–156.
Slany, A., Haudek, V. J., Gundacker, N. C., Griss, J.,
Mohr, T., Wimmer, H., Eisenbauer, M., Elbling, L.,
and Gerner, C. (2009). Introducing a new parame-
ter for quality control of proteome profiles: consider-
ation of commonly expressed proteins. Electrophore-
sis, 30(8):1306–28.
Whistler, T., Rollin, D., and Vernon, S. D. (2007). A method
for improving seldi-tof mass spectrometry data qual-
ity. Proteome Science, 5:14.
Widlak, P., Pietrowska, M., Wojtkiewicz, K., Rutkowski, T.,
Wygoda, A., Marczak, L., Marczyk, M., Polanska, J.,
Walaszczyk, A., Dominczyk, I., Skladowski, K., Sto-
biecki, M., and Polanski, A. (2011). Radiation-related
changes in serum proteome profiles detected by mass
spectrometry in blood of patients treated with radio-
therapy due to larynx cancer. Journal of Radiation
Research, 52(5):575–581.
Wong, J. W., Durante, C., and Cartwright, H. M. (2005).
Application of fast fourier transform cross-correlation
for the alignment of large chromatographic and spec-
tral datasets. Analytical Chemistry, 77(17):5655–61.
Wu, B., Abbott, T., Fishman, D., McMurray, W., Mor,
G., Stone, K., Ward, D., Williams, K., and Zhao, H.
(2006). Ovarian cancer classification based on mass
spectrometry analysis of sera. Cancer Inform, 2:123–
32.
MethodsforQualityControlofLow-resolutionMALDI-ToFSpectra
177
